-

-
left: talk on Data ownership & Collective Awareness in the AI era, given at MYDATA 2019 Helsinki; middle: work; right: relax
-
(Here photos on Cambridge exam "lifestyle")
-
(Here after teaching kids how to code)
- I am Full Professor at the department of Computer Science and
Technology of the University
of Cambridge and I am a member of the Artificial
Intelligence group. I am a member of the
Cambridge Centre for AI in Medicine.
- My research interest focuses on developing Artificial Intelligence and Computational Biology models to understand diseases complexity and address personalised and precision medicine. Current focus is on Graph Neural Network modeling.
- I have a MA from Cambridge, a PhD in
Complex Systems and Non Linear Dynamics (School of
Informatics, dept of Engineering of the University
of
Firenze, Italy) and a PhD in (Theoretical) Genetics (University
of Pavia, Italy).
- Other Affliations: I am member of CAMBRIDGE CENTRE FOR AI IN MEDICINE - the Integrate Cancer Medicine Institute, the committee of MPhil in Computational Biology (Stakeholder Group for the CCBI) , steering committee of Cambridge BIG data, VPH-UK (Virtual Physiological Human), Fellow and member of the Council of Clare Hall College , I am member of Ellis, the European Lab for Learning & Intelligent Systems, I am member of the Academia Europaea; I am listed in www.topitalianscientists.org/Top_italian_scientists_VIA-Academy.aspx
-
Admin: member of Complaint Officer/Examination Review Committee
(Cambridge University); reviewer of 4 MPhils (Newcastle
University), steering committee VPH-UK. - I am happy to receive enquiries for
PhD applications. I have successfully completed the equality and
diversity essentials.
Office: FC20; tel: +44 (0)1223-763604; E_mail: Pietro.Lio at cl.cam.ac.uk
Pietro Liò
Data Integration, cross-modality, evidence synthesis, Machine Learning in medicine
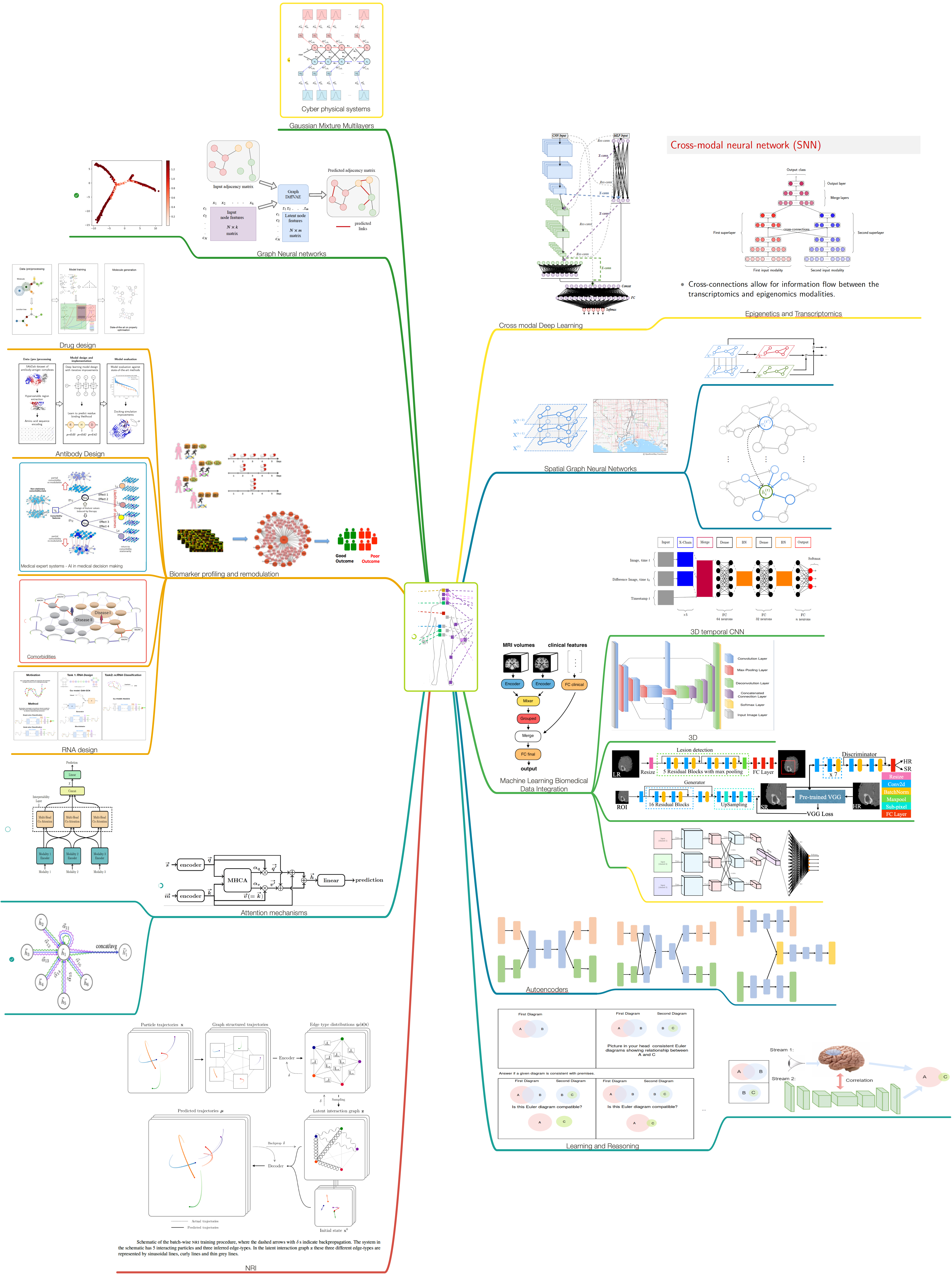
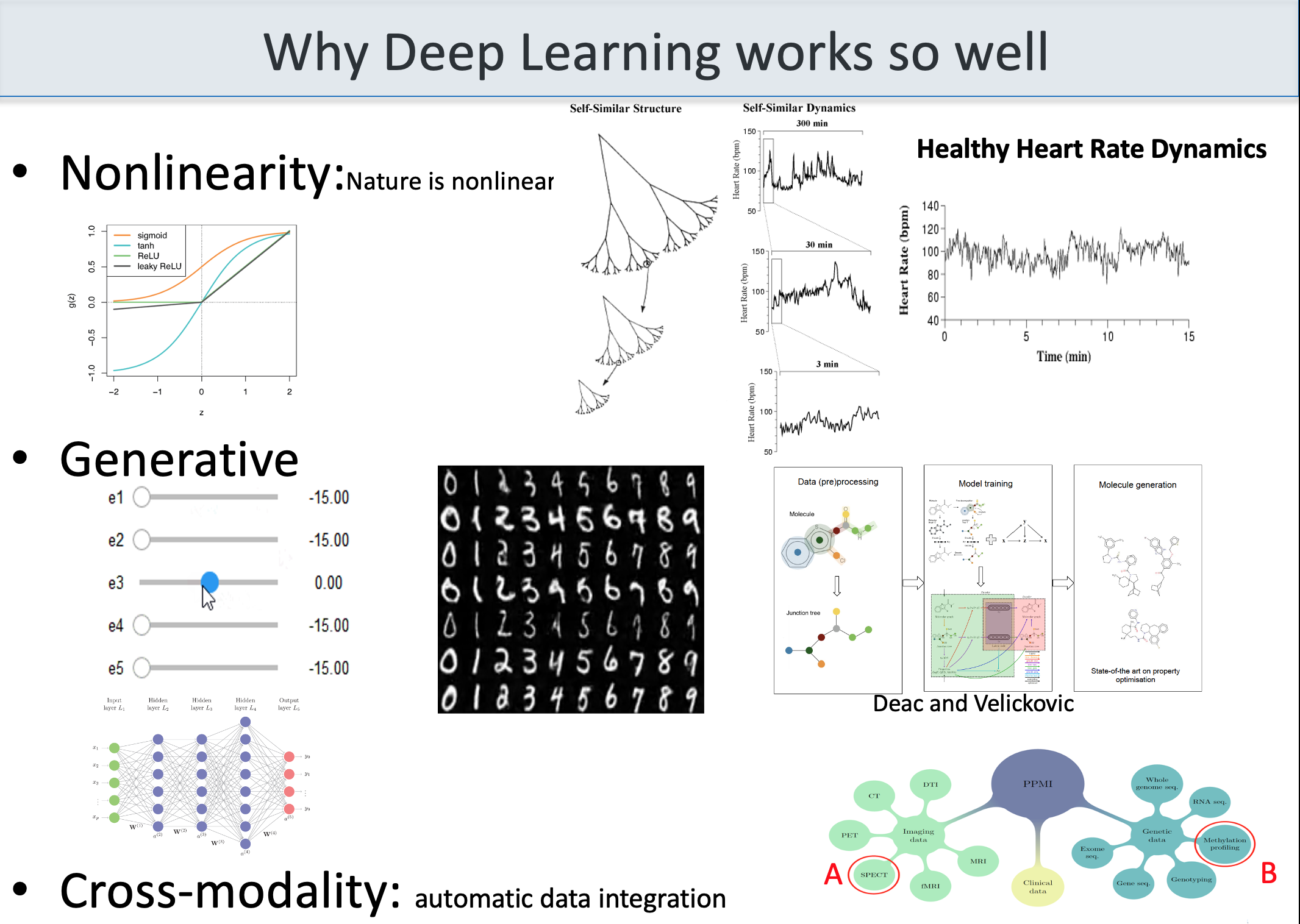
the figure below shows a variety of plots from different papers on deep learning (see at the bottom) focusing on data integration. Data integration is essential to extract all the information, including causality, about a certain subject.
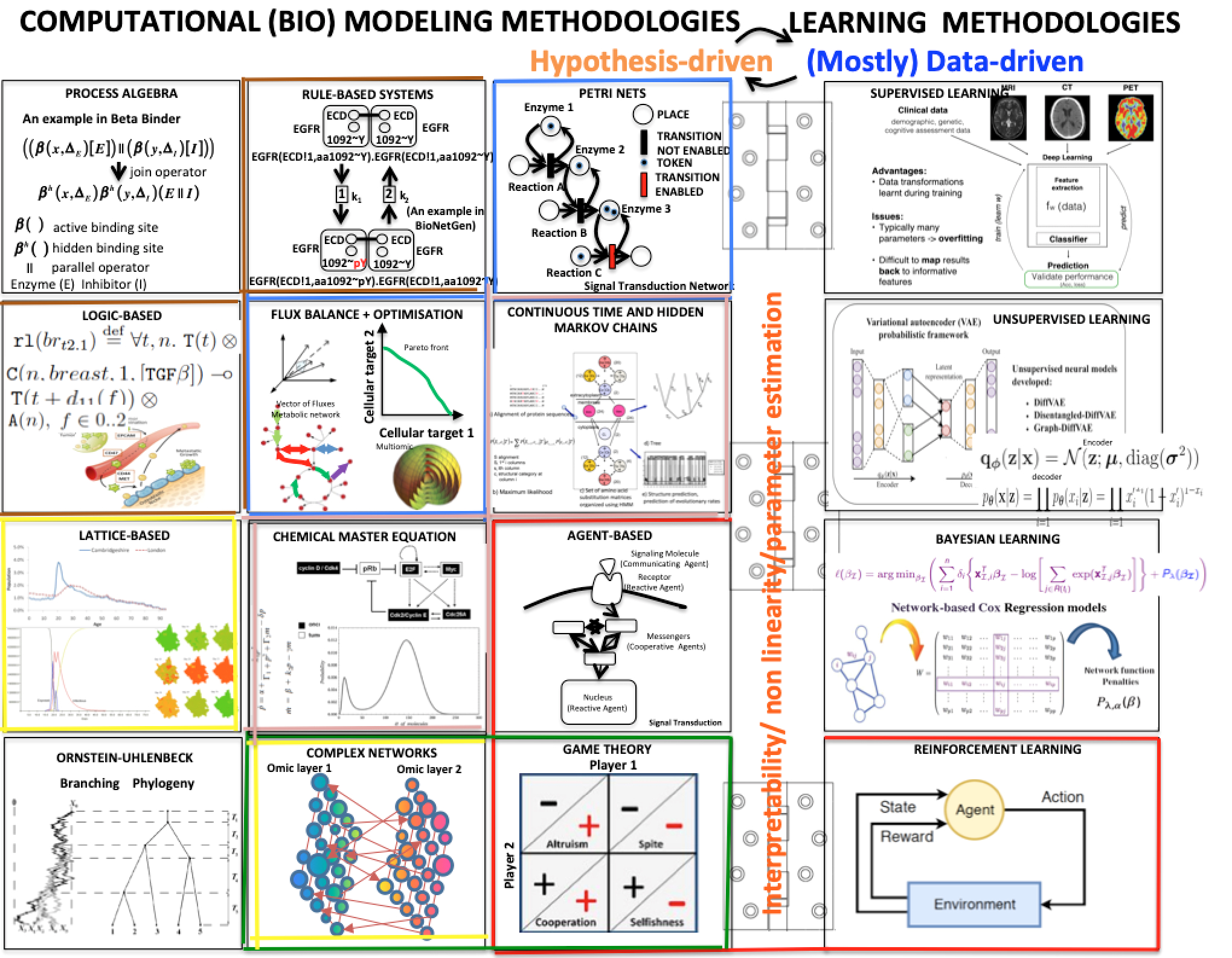
(adapted from Bartocci and Liò, Computational Modeling, Formal Analysis, and Tools for Systems Biology, Plos Computational Biology; also MLCSB 2018: 121-141; NeuroImage 189: 276-287 (2019), CoRR abs/1906.09807 (2019); BMC Bioinformatics 19(1): 439:1-439:18 (2018);CoRR abs/1812.03715 (2018); BMC Bioinformatics 17(S-4): 83 (2016); Mol. BioSyst., 2011, 7, 2796-2803; ACRI 2008: 354-361; Front. Genet., 14 June 2018 | https://doi.org/10.3389/fgene.2018.00206; Bioinformatics 14(8): 726-733 (1998); Fundam. Inform. 171(1-4): 367-392 (2020); BioRxiv, 801605 2019 )
Recent papers, almost full list on Google Scholar or from this pdf
Modeling Social Groups, Policies and Cognitive Behavior in COVID-19 Epidemic Phases. Basic Scenarios (Substantia)
The Computational Patient has Diabetes and a COVID (arxiv)
Forecasting ultra-early intensive care strain from COVID-19 in England (medxiv)
Path Integral Based Convolution and Pooling for Graph Neural Networks
Principal Neighbourhood Aggregation for Graph Nets
Constraining Variational Inference with Geometric Jensen-Shannon Divergence
Other recent papers:
Petar Veličković, William Fedus, William L. Hamilton,
Pietro Lio', Yoshua Bengio, R Devon Hjelm Deep
Graph Infomax. https://arxiv.org/abs/1809.10341
Haider S. at al. Pathway-Based Subnetworks Enable Cross-Disease Biomarker Discovery. In press on Nature Communications.
Editing a Springer Book on Automated Reasoning in Systems Biology and Medicine with Paolo ZulianiJ. Despeyroux, A. Felty, P. Lio’, C. Olarte A Logical Framework for Modelling Breast tumorigenesis. submitted to International Symposium on Molecular Logic and Computational Synthetic Biology.
Hui Xiao, Krzysztof Bartoszek and Pietro Lio'. Multi-omic analysis of signaling factors in inflammatory comorbidities. BMC Bioinformatics (in press)
Ioana Bica, Petar Velickovi ́c, Hui Xiao and Pietro Lio' (2018) Multi-omics data integration using cross-modal neural networks ESANN 2018 proceedings, European Symposium on Artificial Neural Networks, Computational Intelligence and Machine Learning. Bruges (Belgium), 25-27 April 2018
Di Stefano et al, Social Dynamics Modeling of
Chrono-nutrition. Plos Comput. Biology
https://doi.org/10.1371/journal.pcbi.1006714
Edgar Liberis Petar Velickovic, Pietro Sormanni, Michele
Vendruscolo and Pietro Lio' (2018)
Parapred: Antibody Paratope Prediction using Convolutional
and Recurrent Neural Networks. Bioinformatics 2018
Apr 16. doi: 10.1093/bioinformatics/bty305.
Petar Veličković, Guillem Cucurull, Arantxa Casanova,
Adriana Romero, Pietro Lio', Yoshua Bengio Graph
Attention Networks. accepted at ICLR 2018
Scatà M, Di Stefano A, La Corte A, Lio' P. (2018) Quantifying
the propagation of distress and mental disorders in social
networks. Sci Rep. 2018 Mar 22;8(1):5005. doi:
10.1038/s41598-018-23260-2.
Duo Wang, Mateja Jamnik, Pietro Lio' (2018) Investigating
diagrammatic reasoning with deep
neural networks.
Accepted at Diagrams 2018
Bianchi L, Lio' P. Opportunities
for community awareness platforms in personal genomics and
bioinformatics education. Brief Bioinform. 2017 Nov
1;18(6):1082-1090. doi: 10.1093/bib/bbw078
Awards


2016: The paper “Bioaccumulation modelling and sensitivity analysis for discovering key players in contaminated food webs: The case study of PCBs in the Adriatic Sea” (M. Taffi first author) has won the 2016 BYRA first prize at ISEM (The International Society for Ecological Modelling Global Conference) 2016 and the MCED (Modelling Complex Ecological Dynamics) 2016 Award second prize.
2018 - Visiting professor at the University of Padova.
2013 - Lagrange Fellowship (ISI, Universita' Piemonte Orientale).
2012 - Best Paper (Computing with Metabolic Machines) at Turing 100 in Manchester (first author Claudio Angione)
2011 - 3rd prize awarded by the European Commission (sponsored by ERCIM) for the "Methodological bridges for complex systems" (with E. Merelli and N. Paoletti) at the FET 11, 2011 - Future and Emerging Technologies Conference ('Science Beyond Fiction') conference - Budapest
- EVENTS: 20/10/20
talk at TEDxCambridgeUniversity
; 28-30 June 2021: Plenary Lecture at CMBE21 (7th International Conference on Computational and Mathematical Biomedical
Engineering); Politecnico di Milano.
- Group
Members photo1,
photo2, Clare
Hall College December 2019
The post Ph.D. viva of .... try to guess
- My Research
- My Teaching
- Books
- My Impact on Society
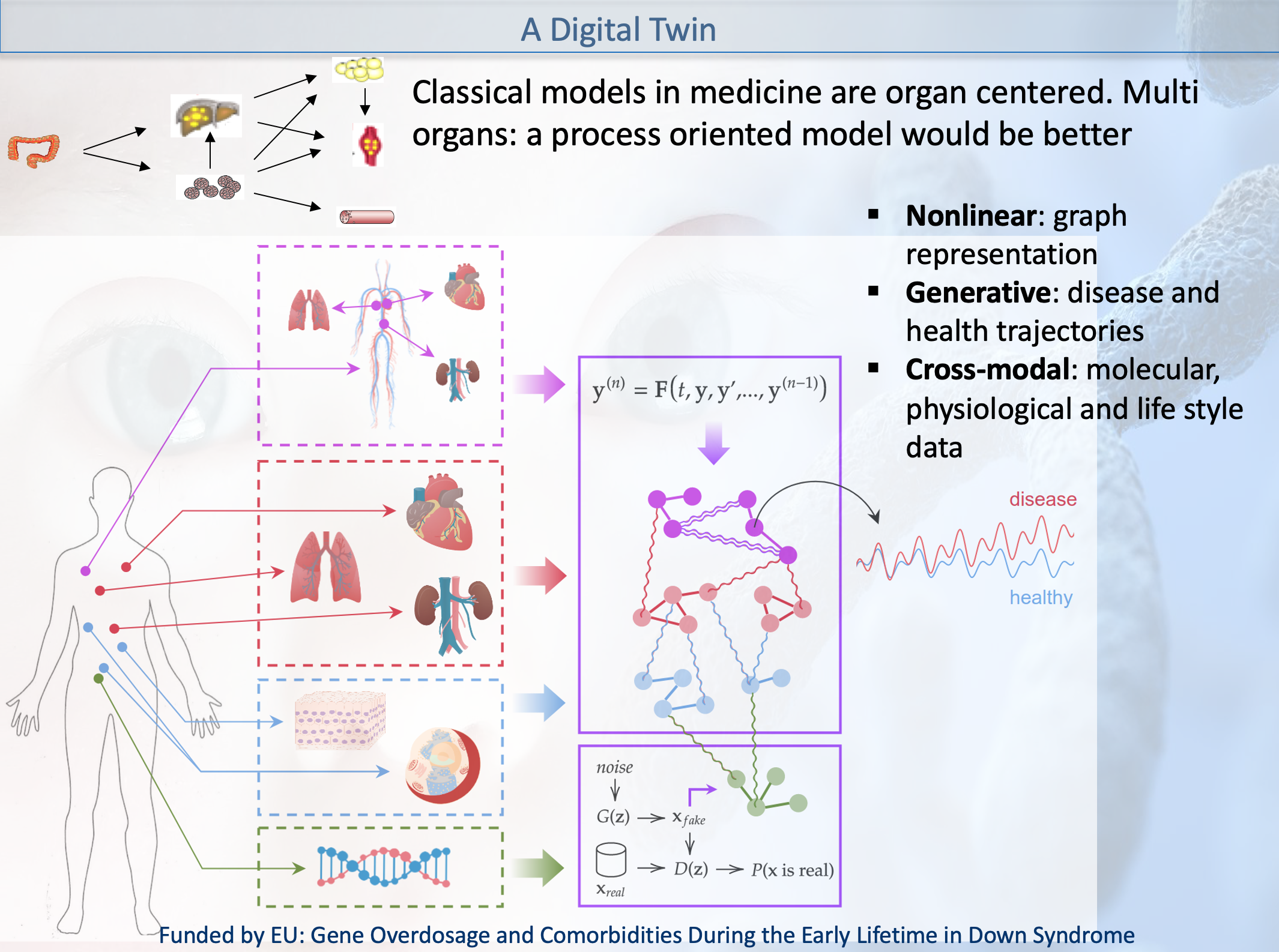
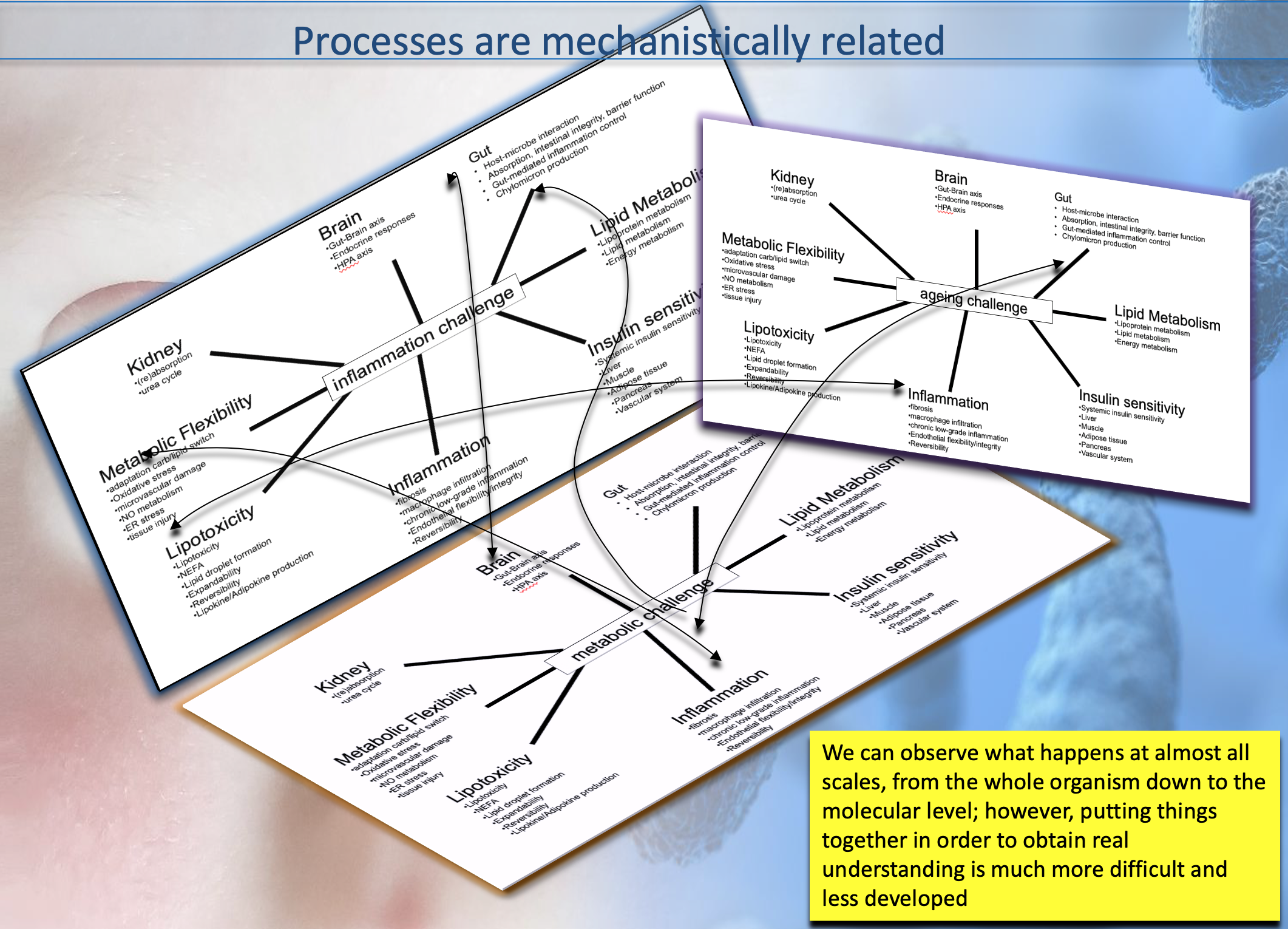
- Examples of past research projects EU Grant: GO-DS21, Gene Overdosage and Comorbidities During the Early Lifetime in Down Syndrome
Currently the group of PhD and postdocs works in three areas: Theory development, Neural signal (fMRI, NMR) analysis, Biomedical methodology development
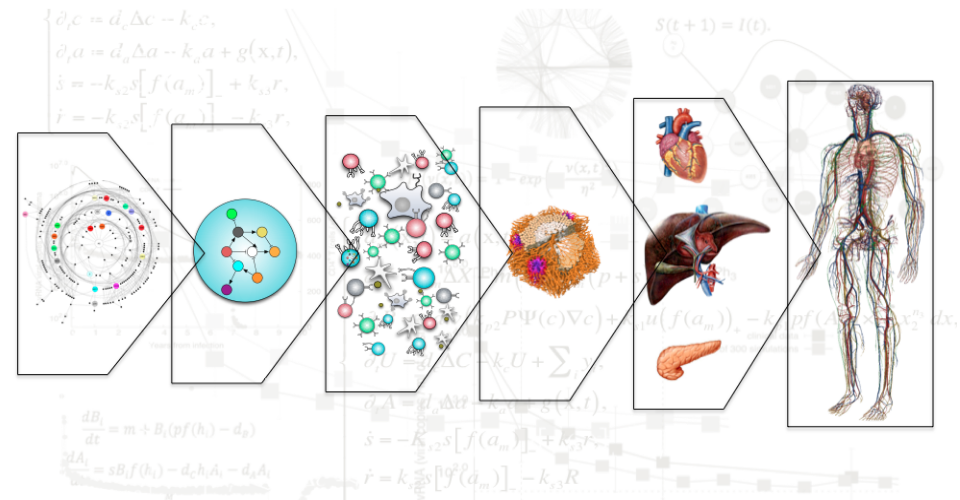
My research consists in developing mechanistic or
phenomenological models across different biological scales
in health and disease conditions; neural networks 'build'
internal (non explainable) models of the system under study.
An important difference between today engineering systems
and biological systems is that Nature is programmed for
self-assembly (i.e. not like IKEA products, more like
self-folding origami). Large sets of reactions involving
molecular complexes of DNA, RNA, proteins, lipids, sugars
etc self assemble into different types of devices
(mechanisms such rotors, motors, control with mechanical and
or electric properties ) that assemble in cells which are
the unit of biology.
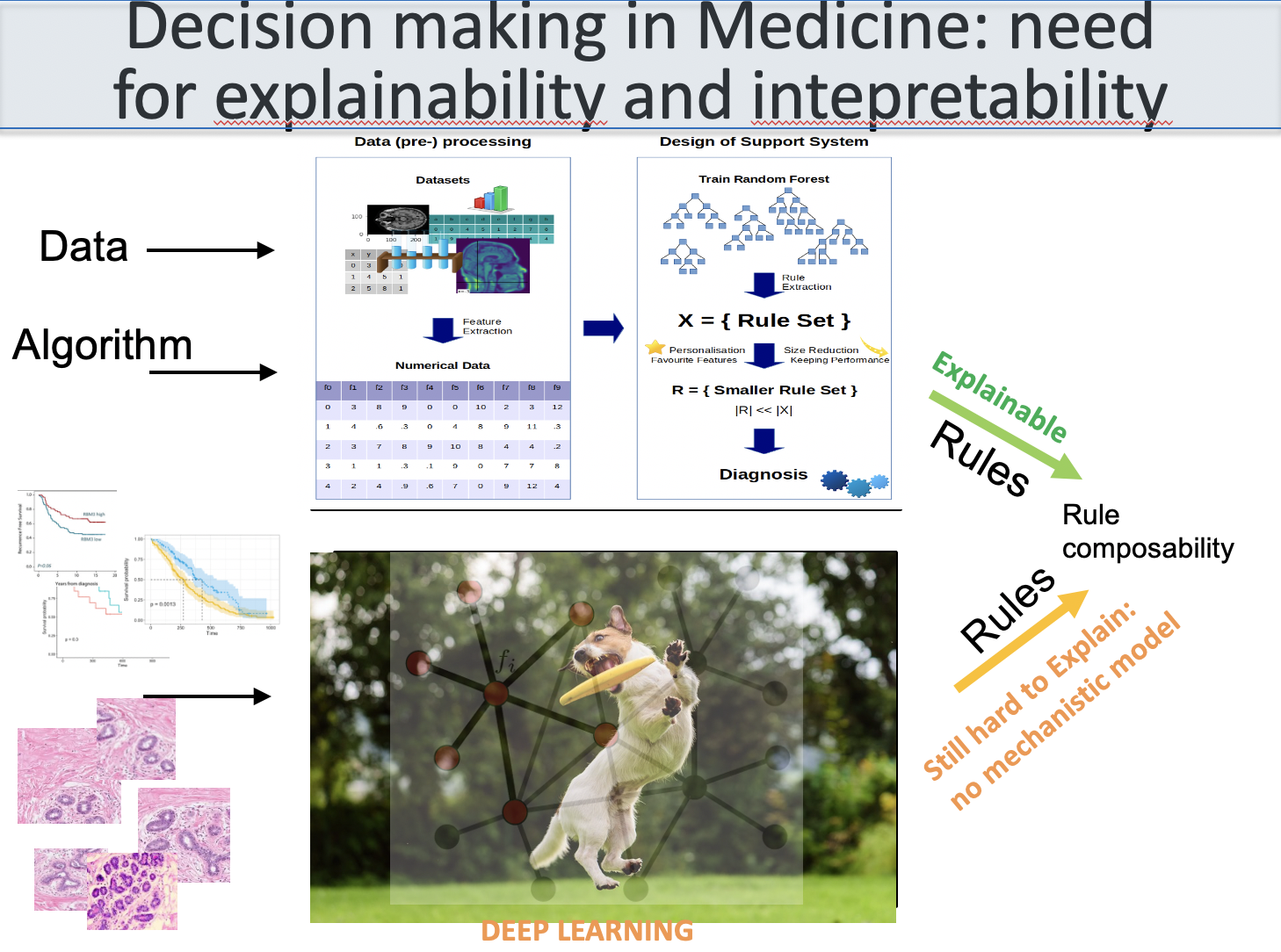
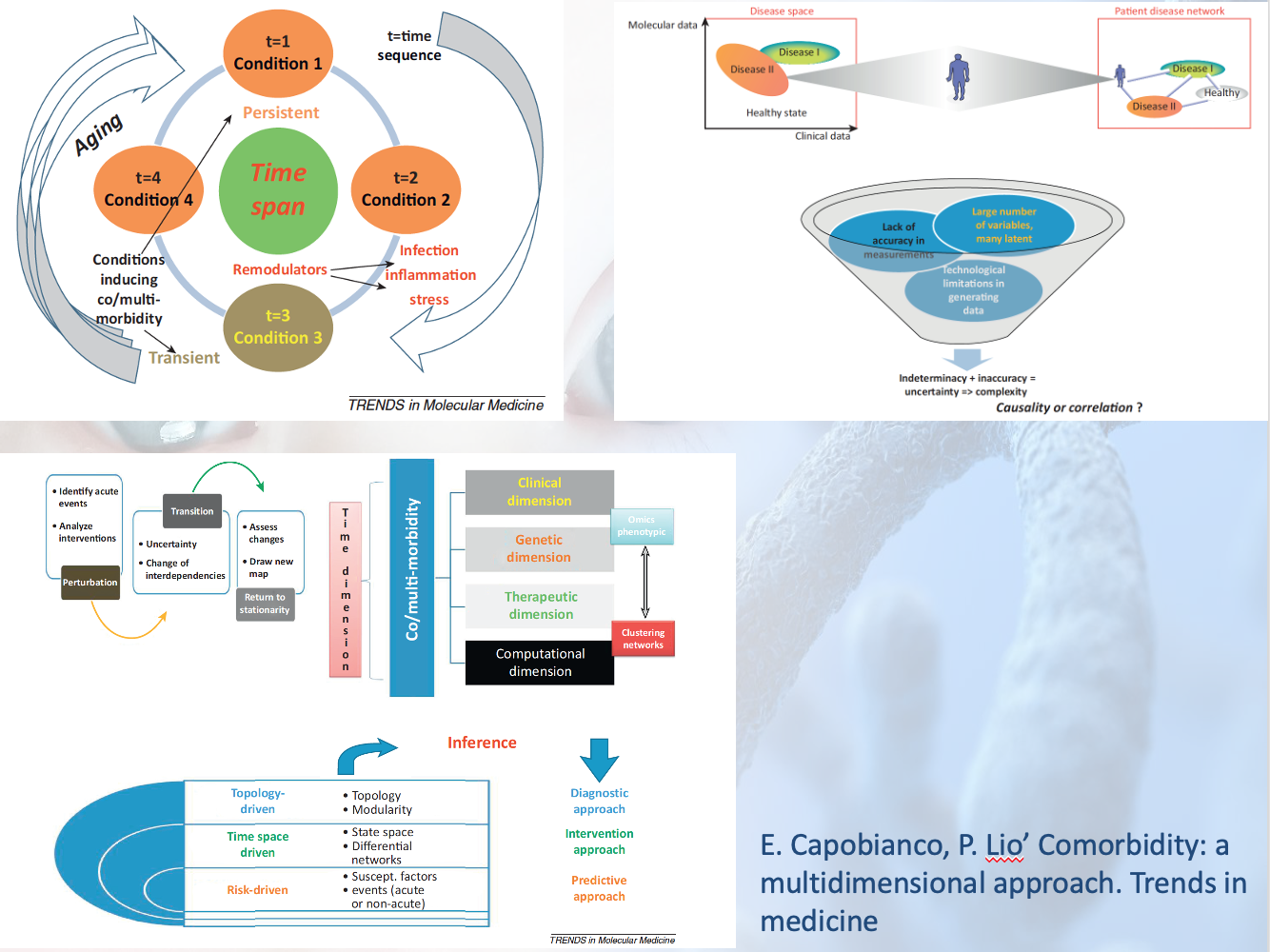
The role of Bioinformatics is (mostly) to identify the
list of parts and their variants. The parts have co-evolved
through mutation and selection events (Nothing in Biology
Makes Sense Except in the Light of Evolution - Theodosius
Dobzhansky). Nowadays, Bioinformatics is mostly based on
algorithms, network science and machine learning. The role
of systems biology is to identify the rules leading to the
multiscale self assembly and produce (dynamical systems or
other) models (Biology is more theoretical than physics -
Bob Lefkowitz's 2012 Nobel lecture) useful for medicine.
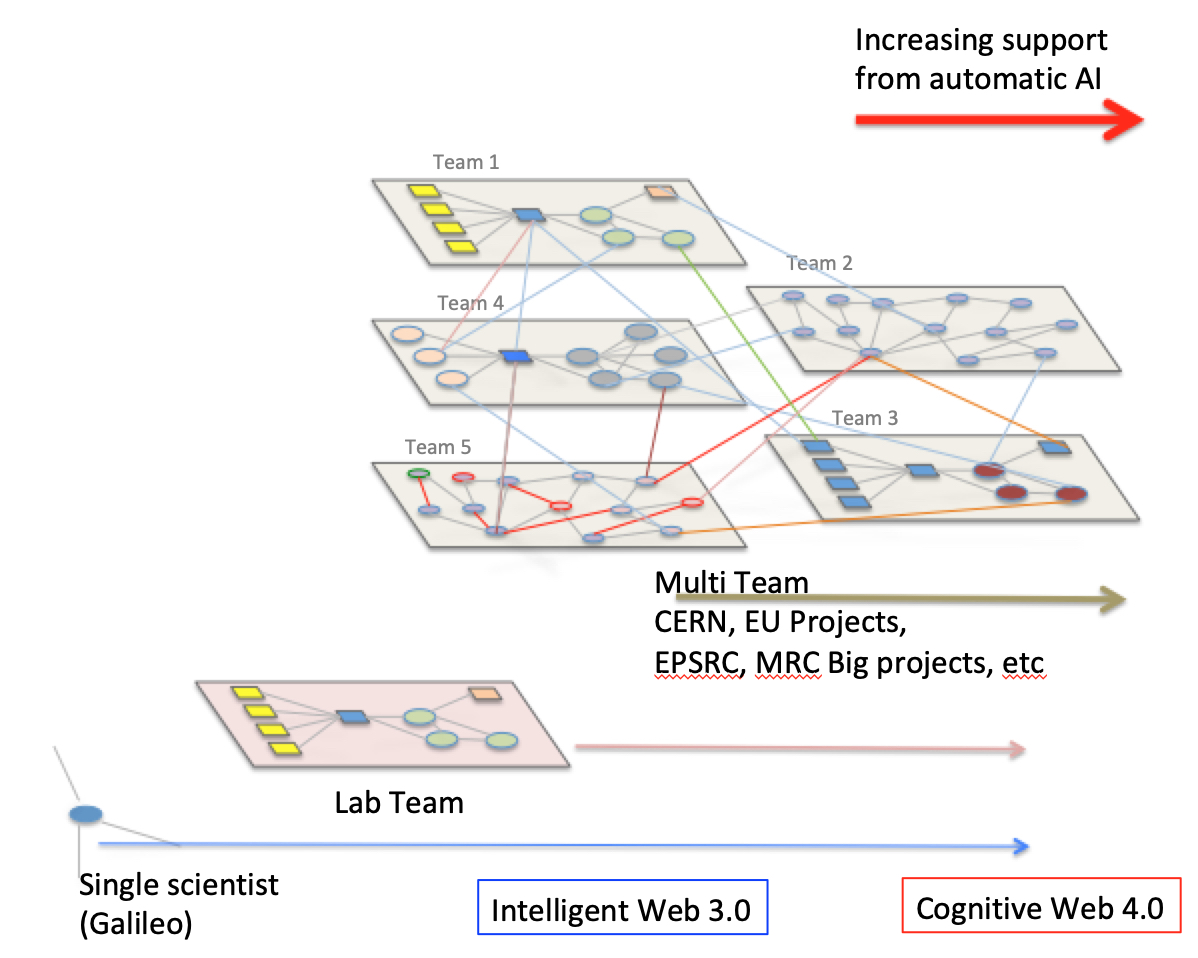
Computational Biology (aka theoretical biology ?) is (in my
opinion) a more general umbrella. Medicine is an evolving
interdisciplinary field (it requires multi-teams work, their
composition depending on the nature of the diagnosed disease
and comorbidities). Computer scientists will play a central
and multi varied role in computational medicine,
particularly addressing challenges of 1) data integration:
genomics and various omics for different tissues, space and
time - such as chromatine, epigenetic, trancriptomic,
proteomic and signalling networks, various microbiota
association with different regions of the body-
epidemiological, electronic health records, radiomics data,
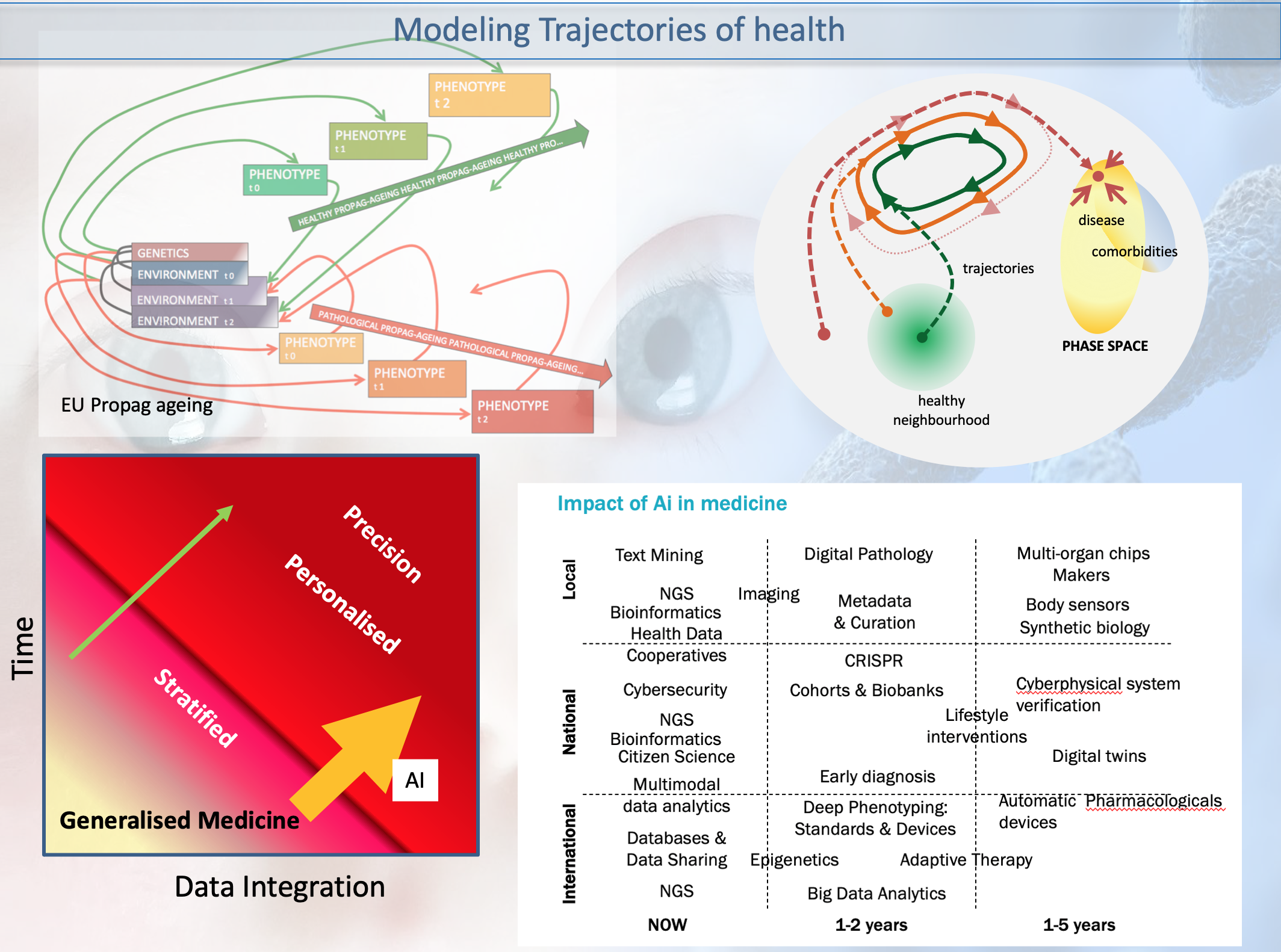
etc and 2) multiscale modelling of the highly nonlinear
disease trajectory -in the Waddington-1968 sense- and taking
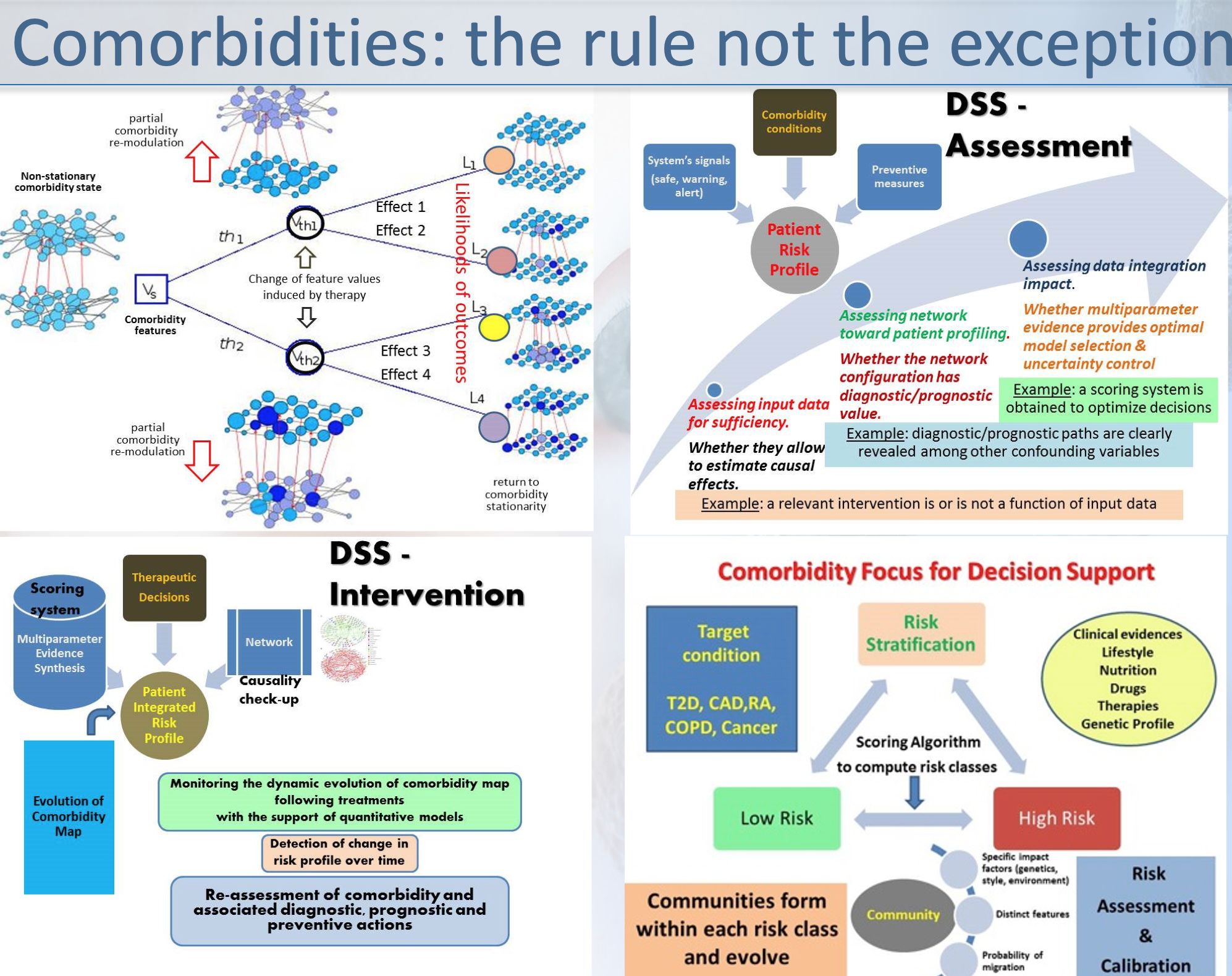
into account biomarkers group remodulation and comorbidity
attractors.
Graph Neural Networks (Advanced Computing MPhil Course).
AI and Biodiversity (AI and Environmental Risk MPhil course)


I am active in science communication (various articles for
magazines, for instance the italian edition of Scientific
American), and engage often with the media and in public
science events (science cafe' in various cities,
participation at European Researchers' Night, Coding for
kids schools, education in computer science such as CS4HS in
2010 (Education in Computer Science for High School
Educators) at Carnegie Mellon University, etc). This is very
important as a scientist is fundamentally a visionary who
needs to interact with society.
I have been involved for several years in the remarkable
EUROPEAN COMMUNITY CAPSSI (Collective Awareness Platforms
for Sustainability and Social Innovation) and NGI (Next
Generation Internet) Initiatives that have pioneered new
models to create awareness of emerging sustainability
challenges (CAPSSI) and inclusivity, openness, protection of
data (NGI).

EU Grant: PROPAG-AGEING - The continuum between healthy ageing and idiopathic Parkinson Disease within a propagation perspective of inflammation and damage: the search for new diagnostic, prognostic and therapeutic targets.

MRC Grant: CLIMB (R. Cardinal) Clinical Informatics for Mind and Brain health.
EU grant: MIMOmics (Methods for Integrated analysis of multiple Omics datasets).
EU grant: Epihealthnet: To improve the health of the human population by understanding mechanisms and pathways in early development, with special emphasis on epigenetic changes and developmentally relevant metabolic signals, which create biological variation and have a long term effect on the health of individuals.
EU Grant: MISSION-T2D aims at developing and validating an integrated, multilevel patient-specific model for the simulation and prediction of metabolic and inflammatory processes in the onset and progress of the type 2 diabetes (T2D).
NERC (Natural Environment Research council) DREAM (Data, Risk and Environmental Analytical Methods): The CDT research underway seeks to utilise emerging technologies, techniques and tools, to more accurately monitor the environment, enabling cutting edge research.
EU Grant Metable:-Advanced bioinformatics for genome and metagenome analyses with discovery of novel biocatalysts from extremophiles: implications for improving industrial bioprocesses.
Socialnets: Social networking for pervasive adaptation