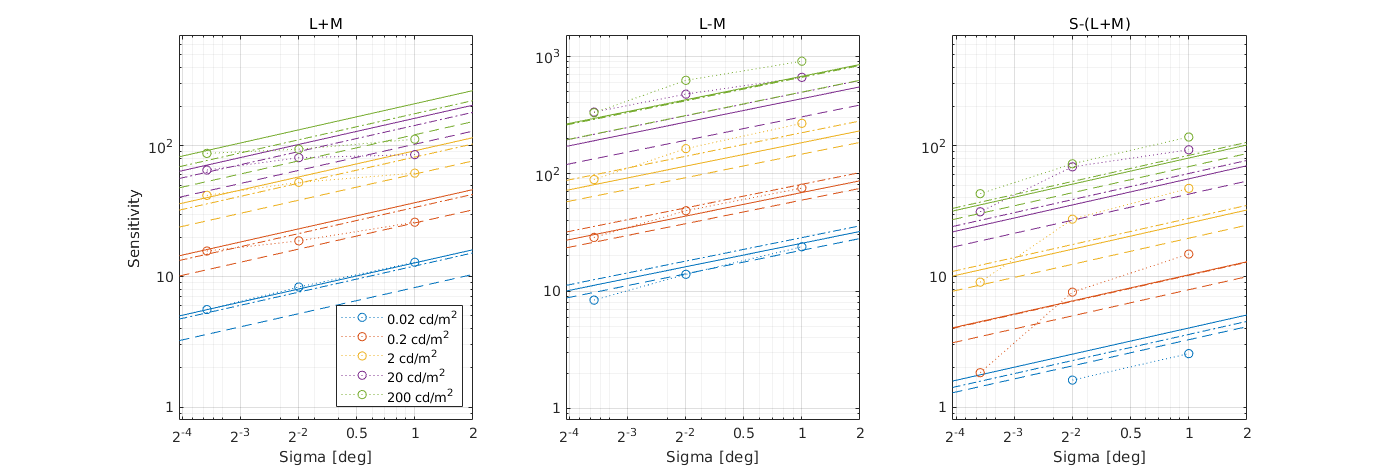
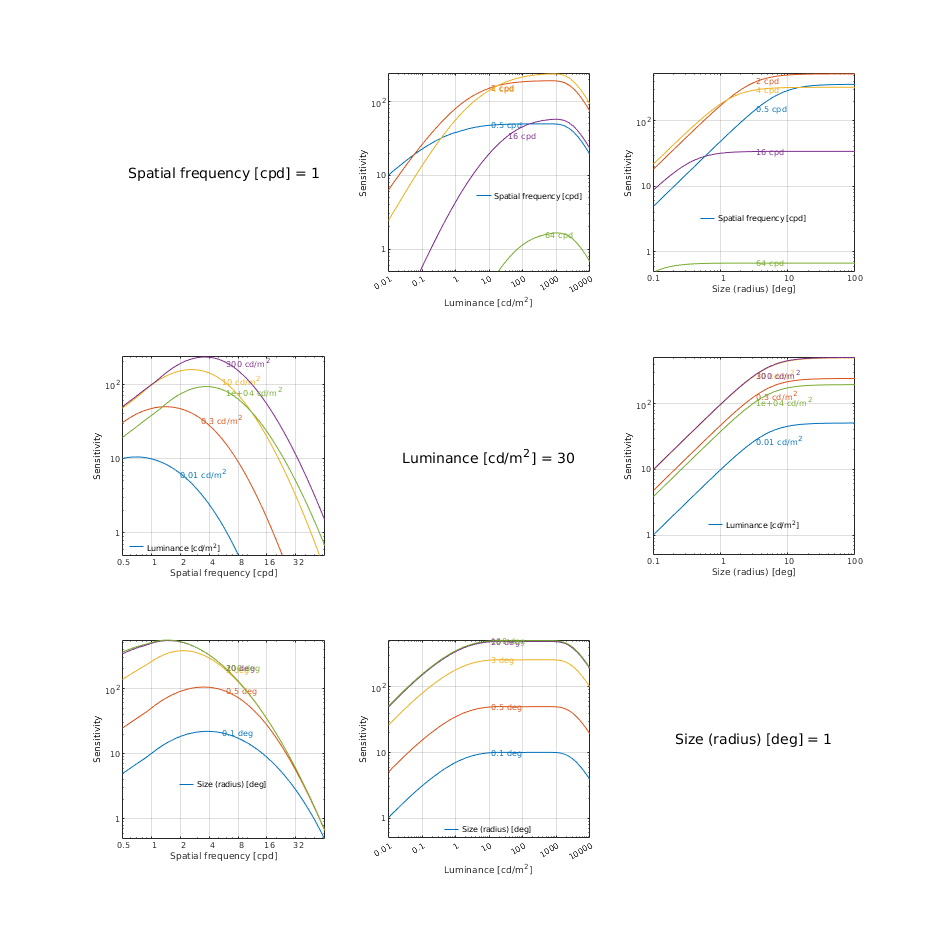
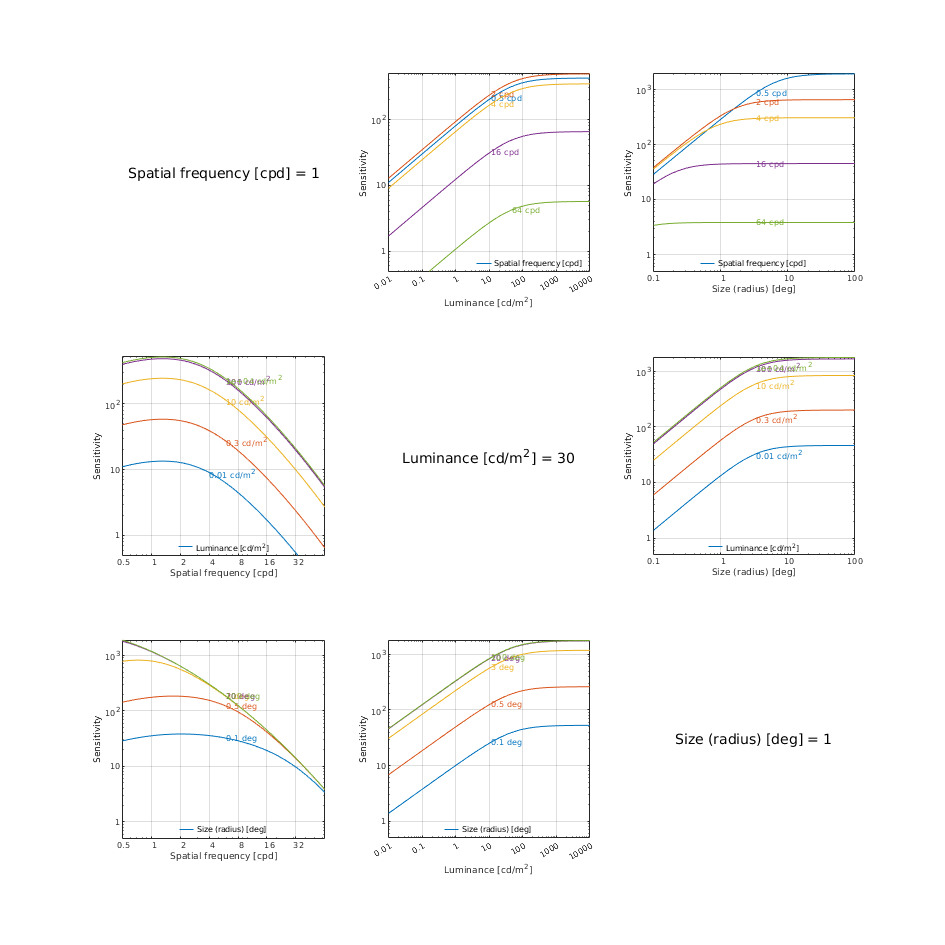
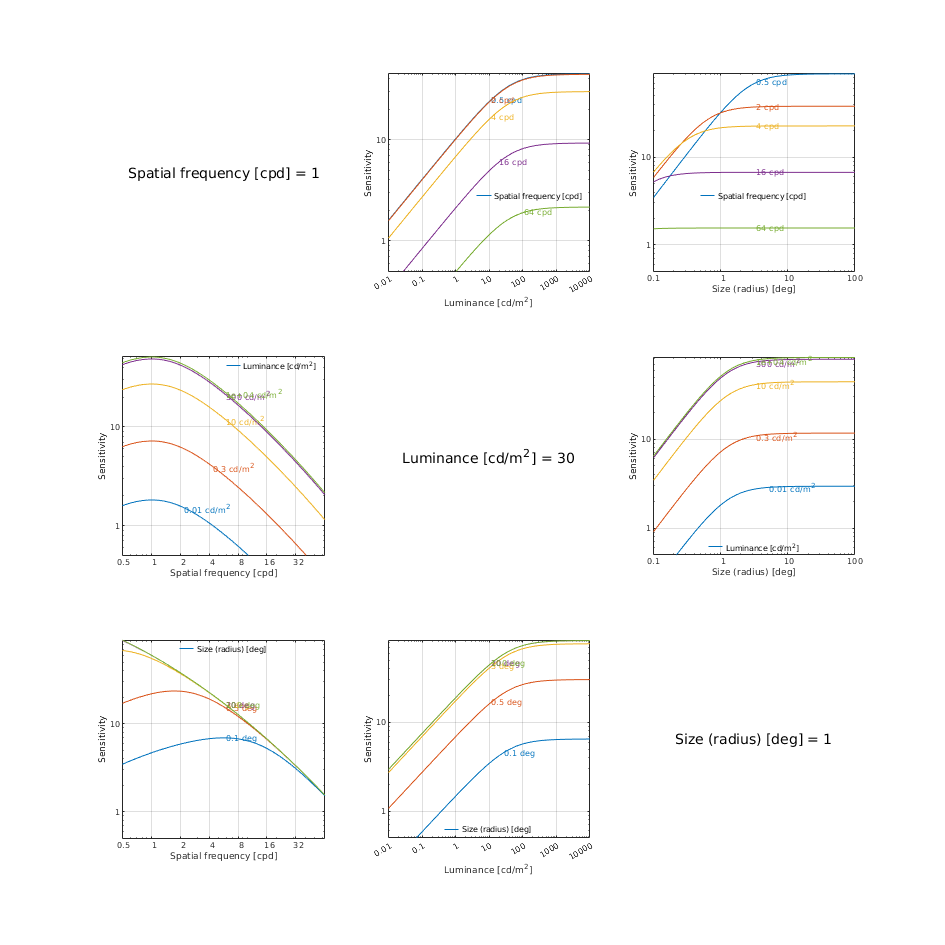
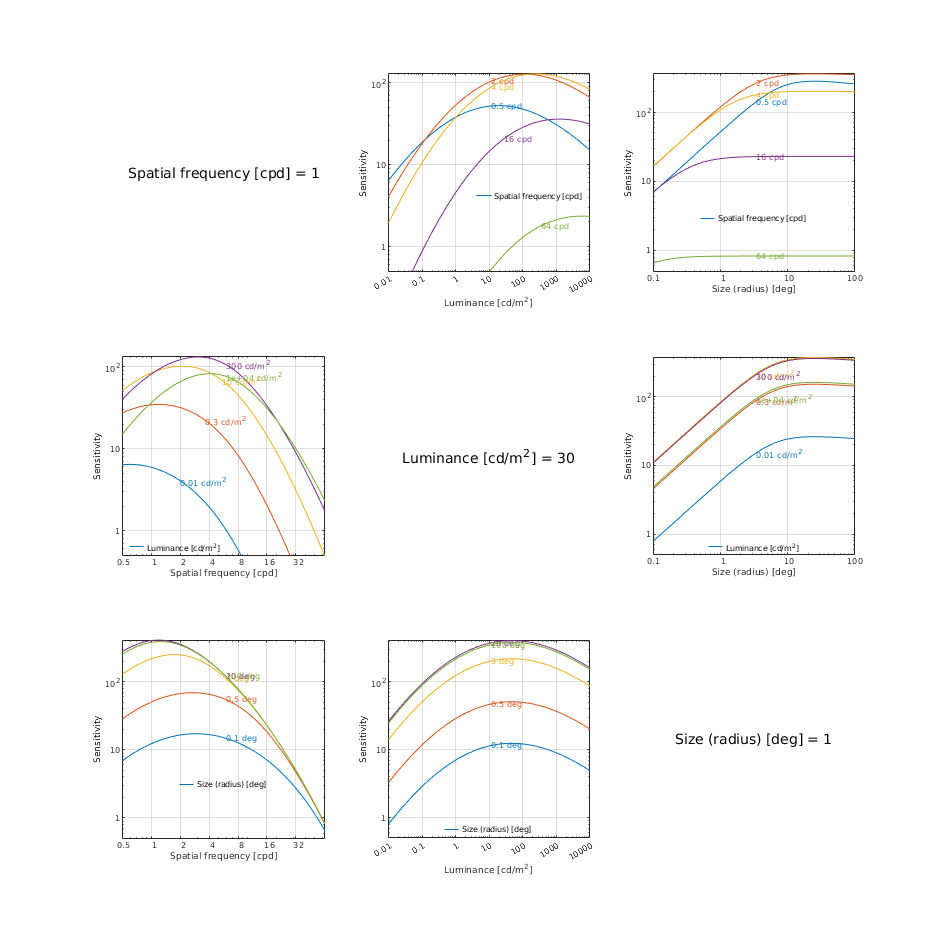
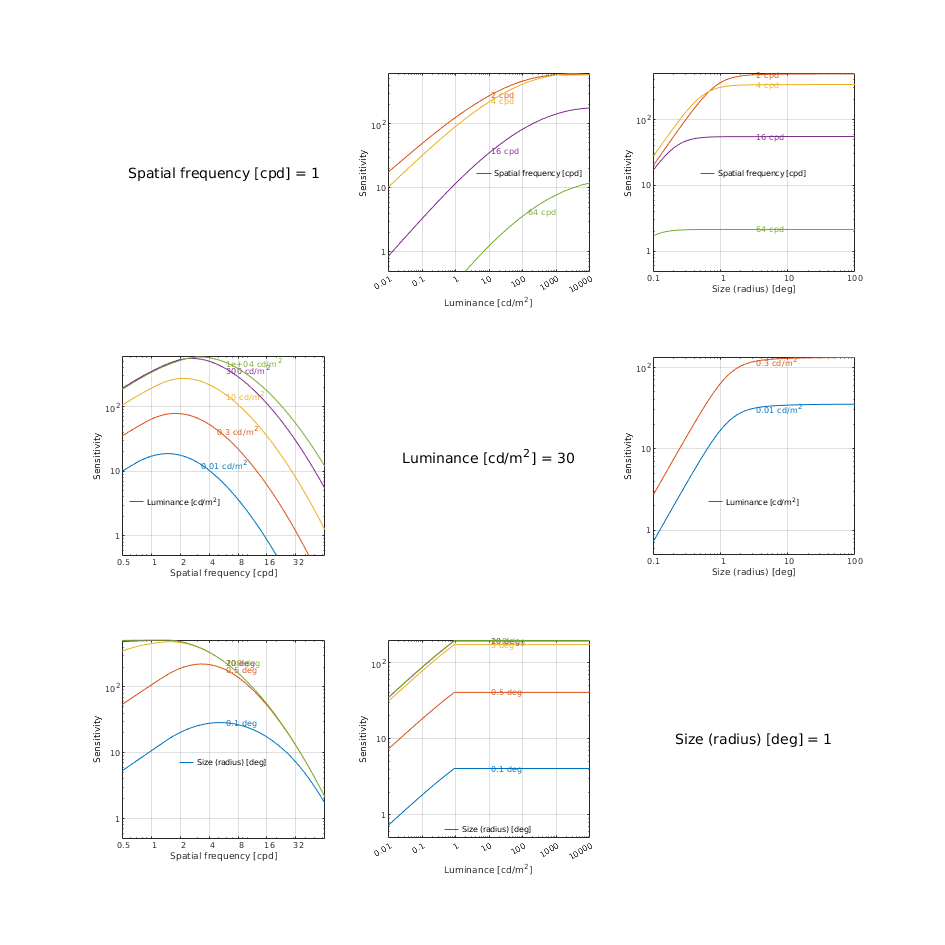
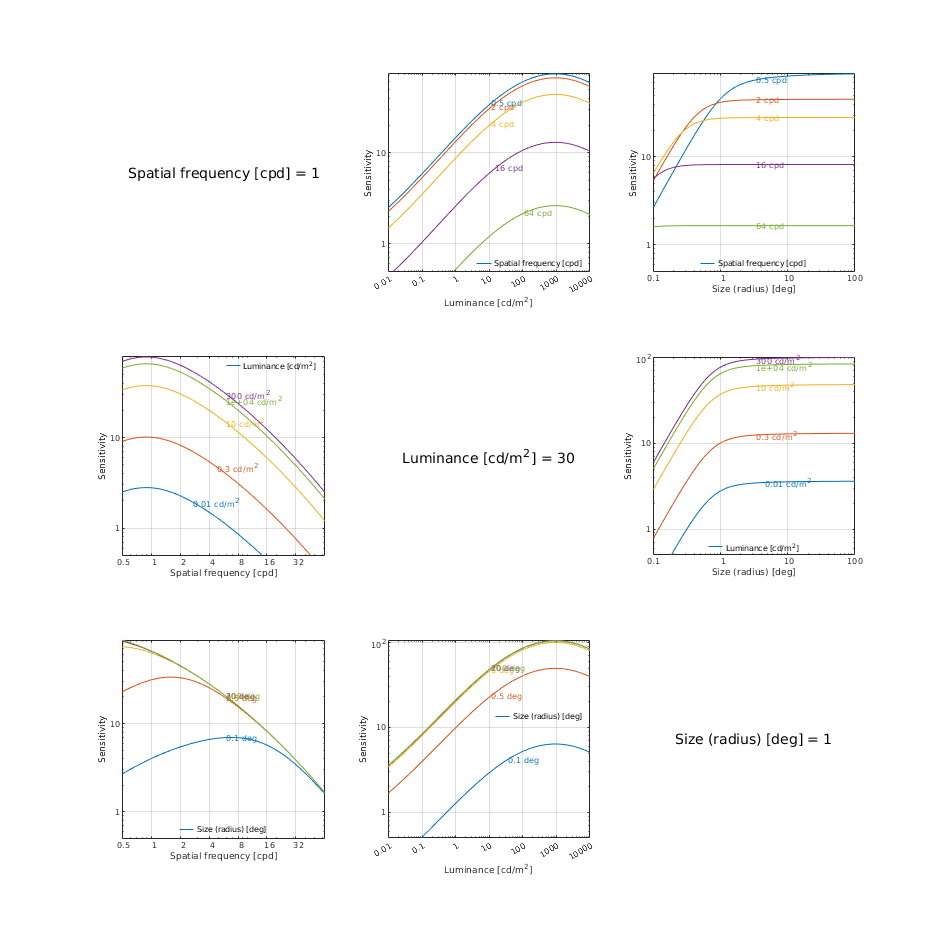
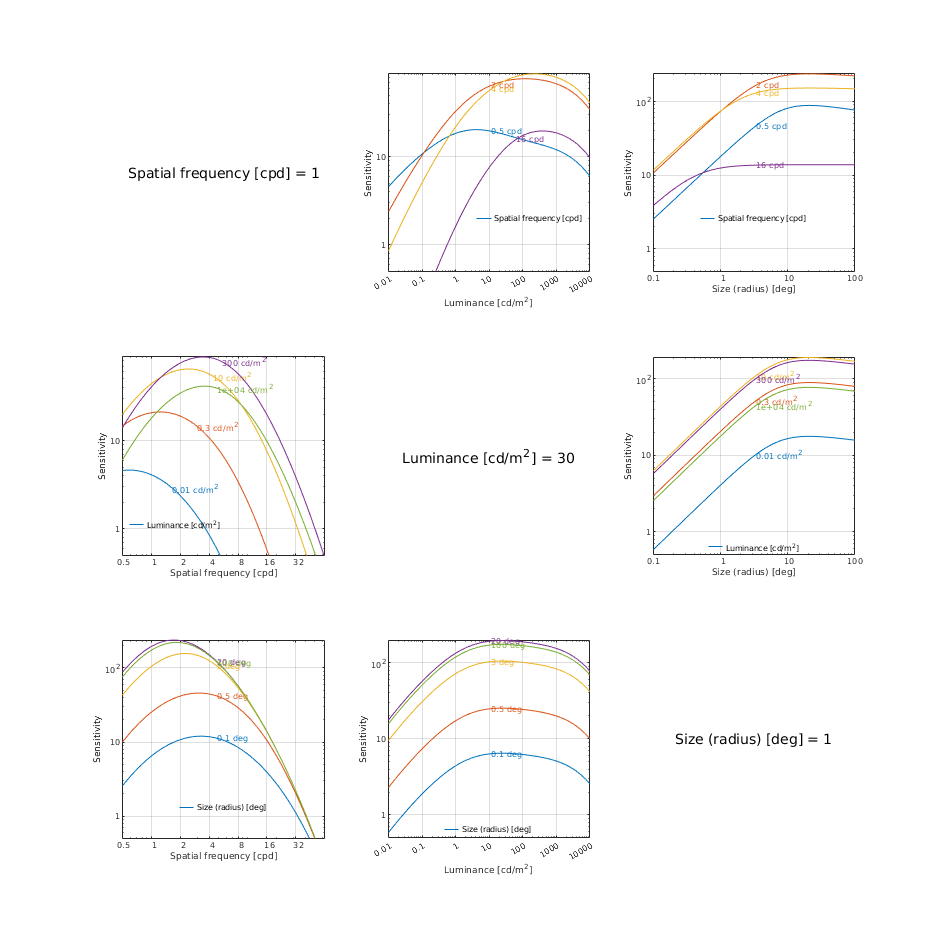
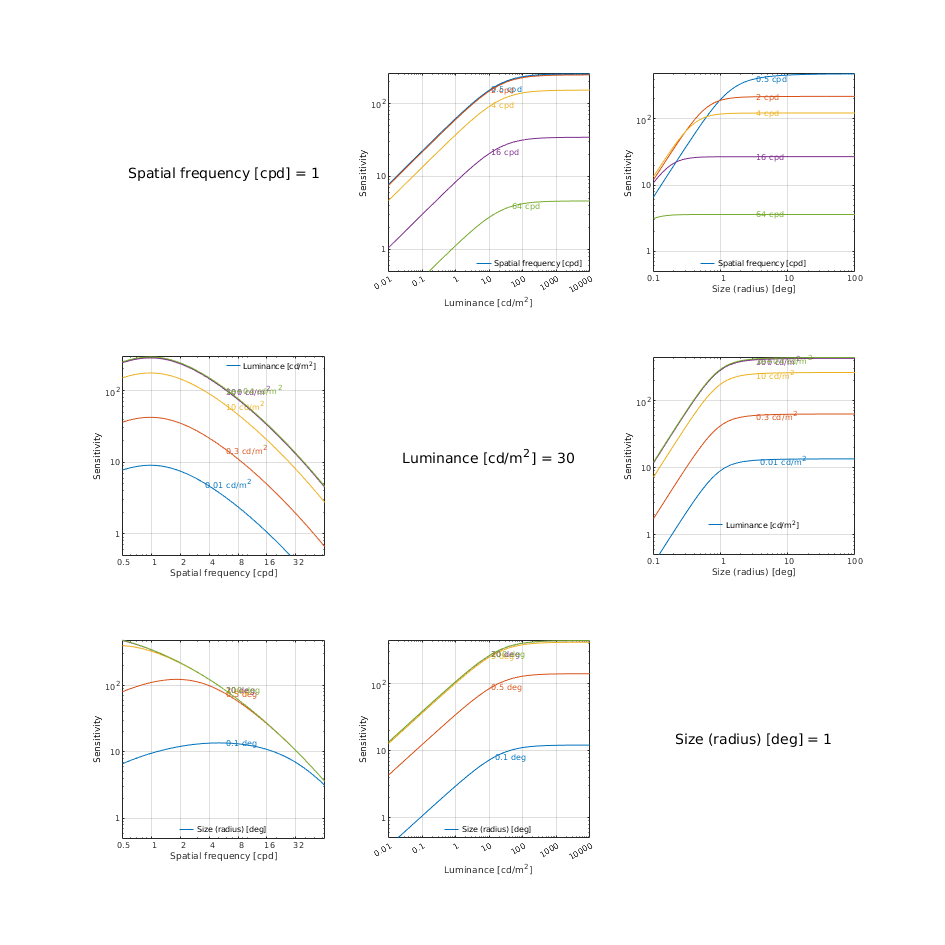
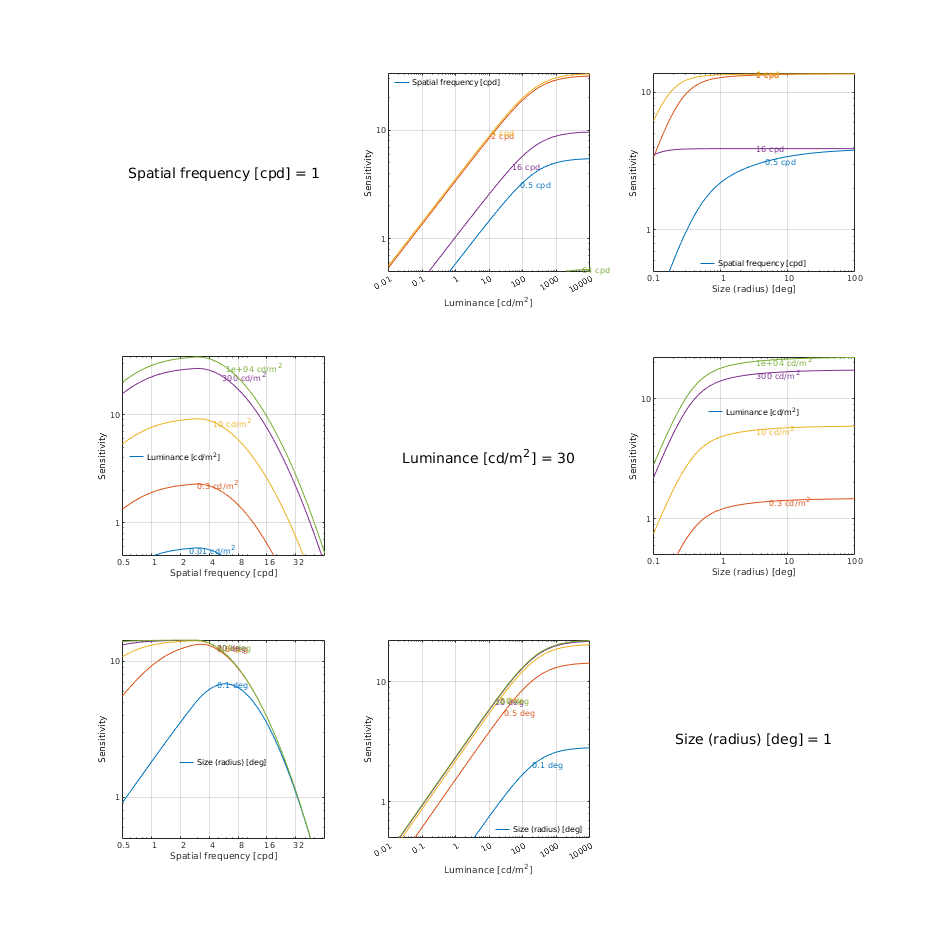
CSF models as functions of spatial, luminance, and area for three DKL colour directions
Fitting error
Model comparison statistics
We use AIC and F-test to test whether the difference in fitting error is statistically significant at alpha=0.05 level. Both statistical metrics take the number of optimized parameters into account.
F-test: For F-test, we compare the fitting results from castleCSF with those of other models. The F-static is calculated using the residual sum of squares and degrees of freedom (number of data points - number of optimized parameters) from both models. The corresponding p-value indicates whether or not the null hypothesis is rejected, where H0: the castleCSF does not provide significant better fit than the other model. The p-values less than 0.05 indicates that castleCSF provides a better fit to the data at the significance level of 0.05 (marked with ✓). We performed the F-test for all individual datasets as well as for all datasets combined. For smaller datasets, where the number of data points are comparable to the number of model parameters, F-test can not provide any results since it indicates there is more variance within the models' fits than between.
AIC: Akaike information criterion is a statistical estimator of prediction error and relative quality of the models, which accounts for the number of parameters of each model.
The model with the lower AIC score is considered to be better and with a good balance of error value and the number of parameters.
The sensitivity adjustment column contains a multiplier that is used to adjust the sensitivity of each datasets. It corresponds to sd in the paper.
Model parameters
castleCSF
M_lms2acc =
1.0000 1.0000 0
1.0000 -2.3112 0
-1.0000 -1.0000 50.9875
p.rg.sigma_sust = 16.541;
p.rg.beta_sust = 1.15549;
p.rg.ch_sust.S_max = [ 573.473 48.6858 0.431962 ];
p.rg.ch_sust.f_max = 0.0607044;
p.rg.ch_sust.bw = 1.61835;
p.rg.A_0 = 3346.39;
p.rg.f_0 = 0.103138;
p.rg.ecc_drop = 0.0591431;
p.rg.ecc_drop_nasal = 2.89648e-05;
p.rg.ecc_drop_f = 2.04986e-69;
p.rg.ecc_drop_f_nasal = 0.180118;
p.yv.sigma_sust = 7.9187;
p.yv.beta_sust = 0.999363;
p.yv.ch_sust.S_max = [ 91.9035 39.4081 0.405745 ];
p.yv.ch_sust.f_max = 0.00812336;
p.yv.ch_sust.bw = 2.76881;
p.yv.A_0 = 2.41161e+07;
p.yv.f_0 = 0.000461893;
p.yv.ecc_drop = 0.00357397;
p.yv.ecc_drop_nasal = 5.85804e-141;
p.yv.ecc_drop_f = 0.0080878;
p.yv.ecc_drop_f_nasal = 0.0147658;
p.ach.ach_sust.S_max = [ 58.5306 2.13583 0.255781 5.08129e-07 1.00467e+10 ];
p.ach.ach_sust.f_max = [ 1.61977 86.1728 0.219733 ];
p.ach.ach_sust.bw = 0.000212269;
p.ach.ach_sust.a = 0.0309308;
p.ach.ach_sust.A_0 = 157.103;
p.ach.ach_sust.f_0 = 0.702338;
p.ach.ach_trans.S_max = [ 0.193376 2794.72 ];
p.ach.ach_trans.f_max = 0.000327166;
p.ach.ach_trans.bw = 2.70028;
p.ach.ach_trans.a = 0.000241177;
p.ach.ach_trans.A_0 = 3.58581;
p.ach.ach_trans.f_0 = 2.94996;
p.ach.sigma_trans = 0.0858074;
p.ach.sigma_sust = 10.467;
p.ach.omega_trans_sl = 2.38693;
p.ach.omega_trans_c = 4.5616;
p.ach.ecc_drop = 0.0259781;
p.ach.ecc_drop_nasal = 0.0452708;
p.ach.ecc_drop_f = 0.0217926;
p.ach.ecc_drop_f_nasal = 0.0068348;
Parameters for Ach component:
p.ach_sust.S_max = [ 58.5306 2.13583 0.255781 5.08129e-07 1.00467e+10 ];
p.ach_sust.f_max = [ 1.61977 86.1728 0.219733 ];
p.ach_sust.bw = 0.000212269;
p.ach_sust.a = 0.0309308;
p.ach_trans.S_max = [ 0.193376 2794.72 ];
p.ach_trans.f_max = 0.000327166;
p.ach_trans.bw = 2.70028;
p.ach_trans.a = 0.000241177;
p.ach_trans.A_0 = 3.58581;
p.ach_trans.f_0 = 2.94996;
p.sigma_trans = 0.0858074;
p.sigma_sust = 10.467;
p.omega_trans_sl = 2.38693;
p.omega_trans_c = 4.5616;
p.ecc_drop = 0.0259781;
p.ecc_drop_nasal = 0.0452708;
p.ecc_drop_f = 0.0217926;
p.ecc_drop_f_nasal = 0.0068348;
Parameters for RG component:
p.ch_sust.S_max = [ 573.473 48.6858 0.431962 ];
p.ch_sust.f_max = 0.0607044;
p.ch_sust.bw = 1.61835;
p.A_0 = 3346.39;
p.f_0 = 0.103138;
p.sigma_sust = 16.541;
p.beta_sust = 1.15549;
p.ecc_drop = 0.0591431;
p.ecc_drop_nasal = 2.89648e-05;
p.ecc_drop_f = 2.04986e-69;
p.ecc_drop_f_nasal = 0.180118;
Parameters for YV component:
p.ch_sust.S_max = [ 91.9035 39.4081 0.405745 ];
p.ch_sust.f_max = 0.00812336;
p.ch_sust.bw = 2.76881;
p.A_0 = 2.41161e+07;
p.f_0 = 0.000461893;
p.sigma_sust = 7.9187;
p.beta_sust = 0.999363;
p.ecc_drop = 0.00357397;
p.ecc_drop_nasal = 5.85804e-141;
p.ecc_drop_f = 0.0080878;
p.ecc_drop_f_nasal = 0.0147658;
Wuerger 2020 JOV CSF
p.ach.S_max = [ 2.83912 1.28908 43.3254 ];
p.ach.f_max = [ 2.53012 9.8288 44.2978 ];
p.ach.bw = 1.57304;
p.ach.Ac_prime = 115.858;
p.ach.f_0 = 0.44229;
p.ach.gamma = 0.879593;
p.rg.S_max = [ 2.8927 3.17295 43.7516 ];
p.rg.f_max = [ 0.0911927 3.04969e-24 ];
p.rg.bw = 1.2271;
p.rg.Ac_prime = 14.9649;
p.rg.f_0 = 0.781478;
p.rg.gamma = 1.45561;
p.yv.S_max = [ 2.29543 2.99149 24.2307 ];
p.yv.f_max = 0.0788081;
p.yv.bw = 3.17419;
p.yv.Ac_prime = 2.27589;
p.yv.f_0 = 0.330337;
p.yv.gamma = 1.38367;
Postreceptoral contrast
p.ach.S_max = [ 875.389 1.84216 0.237457 15332.4 0.564498 ];
p.ach.f_max = [ 1.38549 80.5781 0.223281 ];
p.ach.bw = 8.75326e-08;
p.ach.gamma = 0.848849;
p.ach.Ac_prime = 157.547;
p.rg.S_max = [ 784.005 22.1349 0.455058 ];
p.rg.f_max = 0.107981;
p.rg.bw = 1.76928;
p.rg.gamma = 1.583;
p.rg.Ac_prime = 5.44558;
p.yv.S_max = [ 955.02 278.691 0.401103 ];
p.yv.f_max = 2.83005;
p.yv.bw = 0;
p.yv.gamma = 1.37576;
p.yv.Ac_prime = 0.502274;
p.colmat = [ 0.116375 1.38979 3.5954 1.05972e-08 3.87096e-06 7.11525 ];
↸CSF model: castleCSF
↸CSF model: Wuerger 2020 JOV CSF
↸CSF model: Postreceptoral contrast
Legend
To keep the plots legible, only up to 3 models are plotted.

↸Dataset: [modelfest] ModelFest
Achromatic CSF as a function of frequency
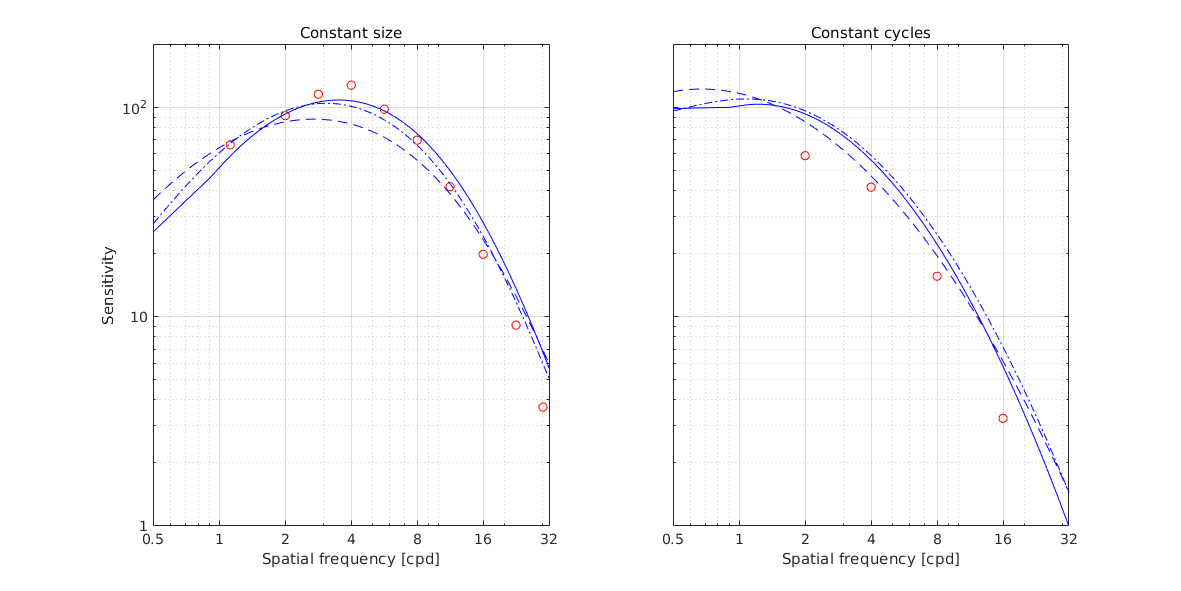
↸Dataset: [hdrvdp_csf] HDR-VDP CSF
Achromatic CSF as a function of frequency
Achromatic CSF as a function of size
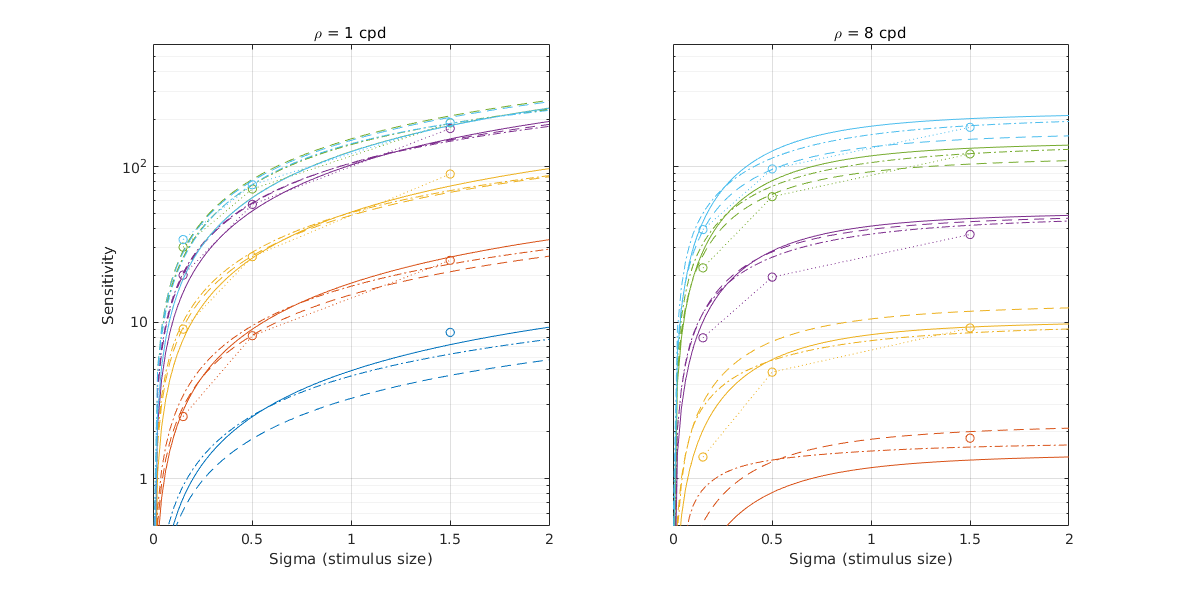
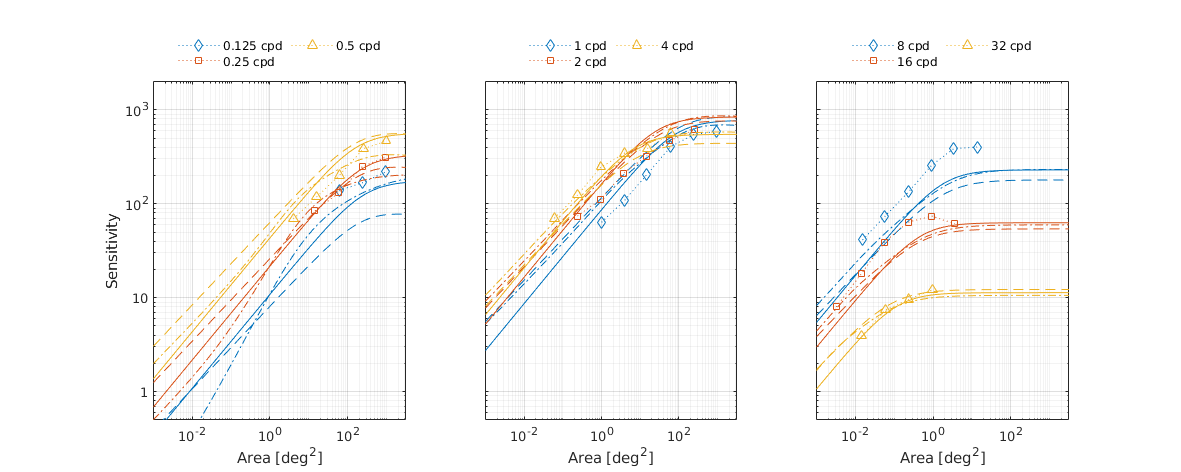
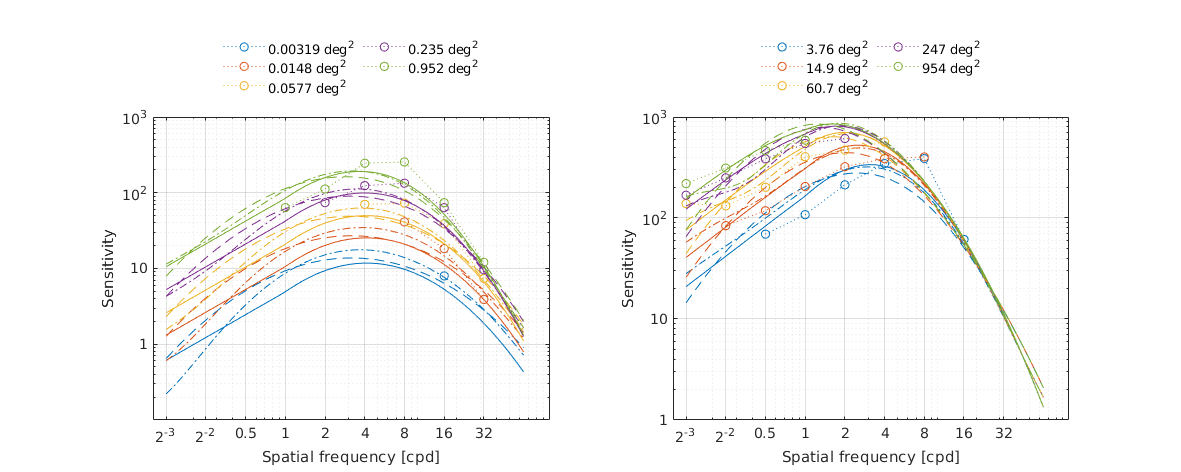
↸Dataset: [rovamo1993] Rovamo et al. 1993
CSF as the funcation of stimulus area
CSF as the function of spatial frequency
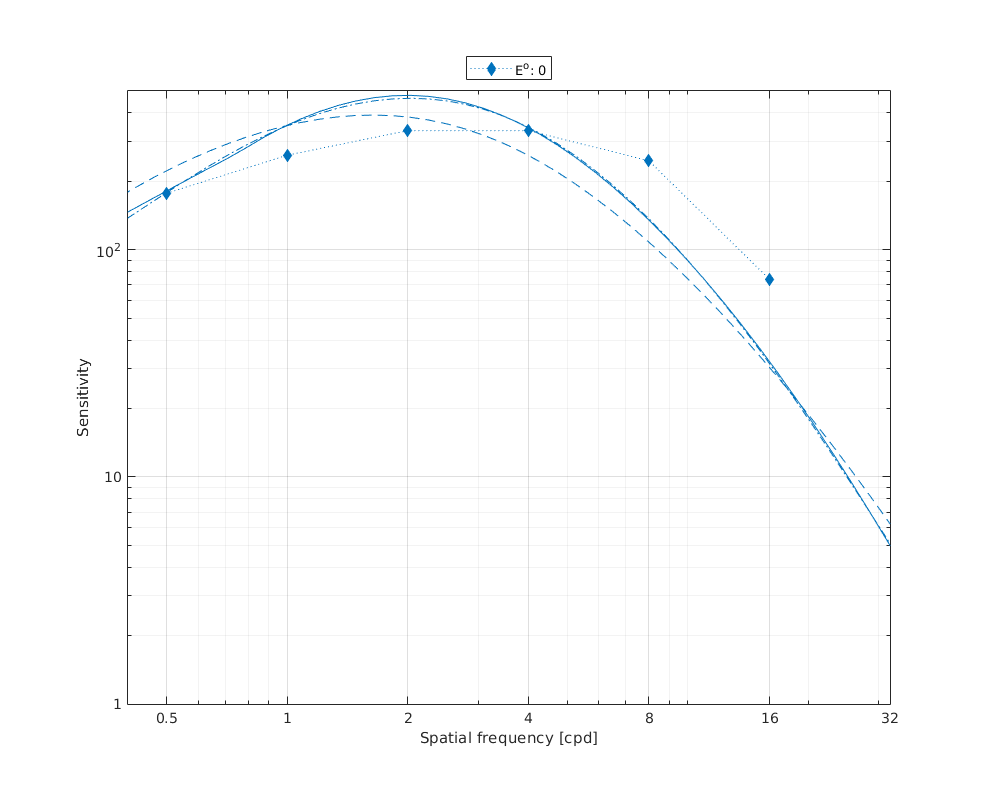
↸Dataset: [virsu1979] Virsu & Rovamo 1979
Contrast sensitivity of central and peripheral vision as a function of spatial frequency and eccentricity
↸Dataset: [wright1983] Wright and Johnson 1983
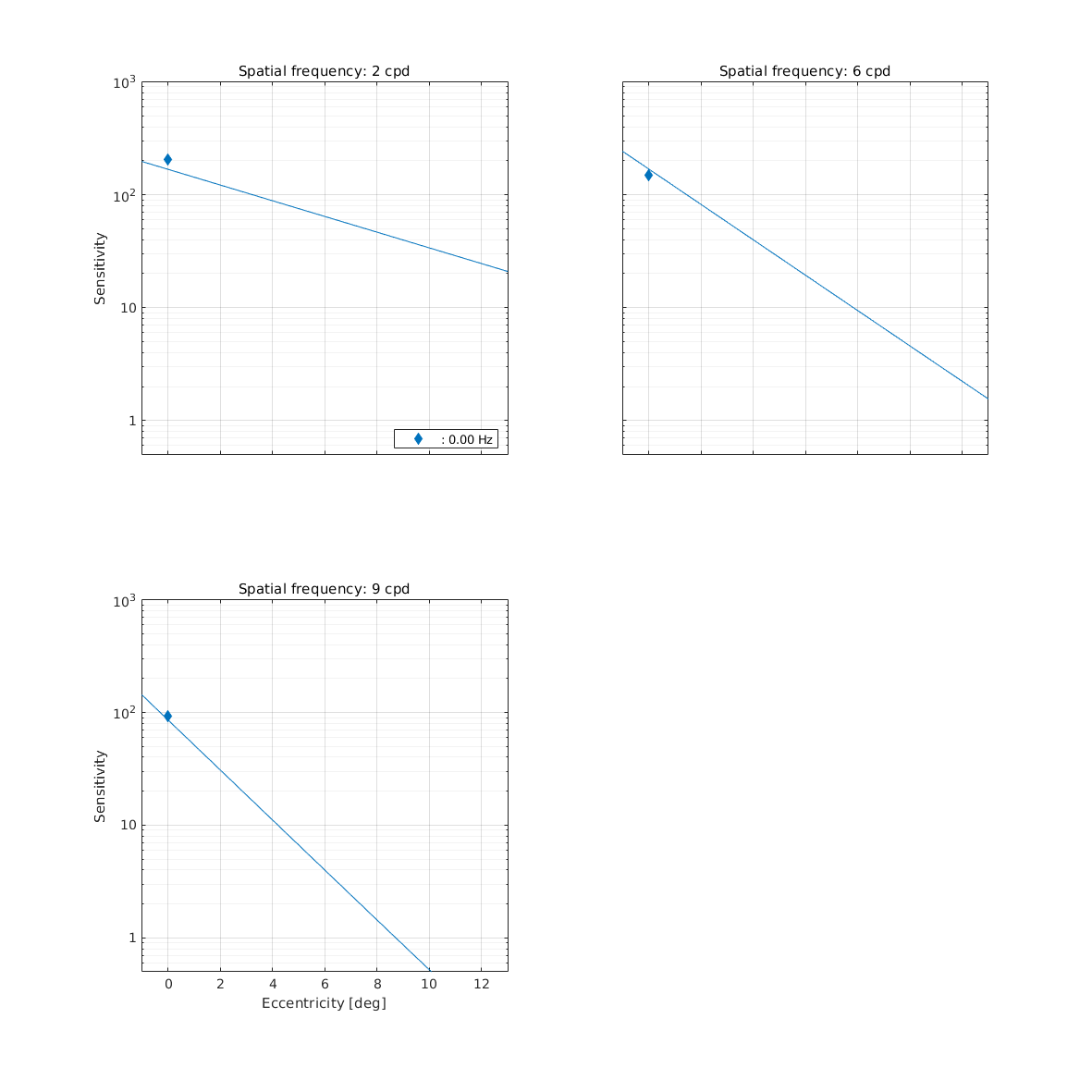
CSF as function of eccentricity
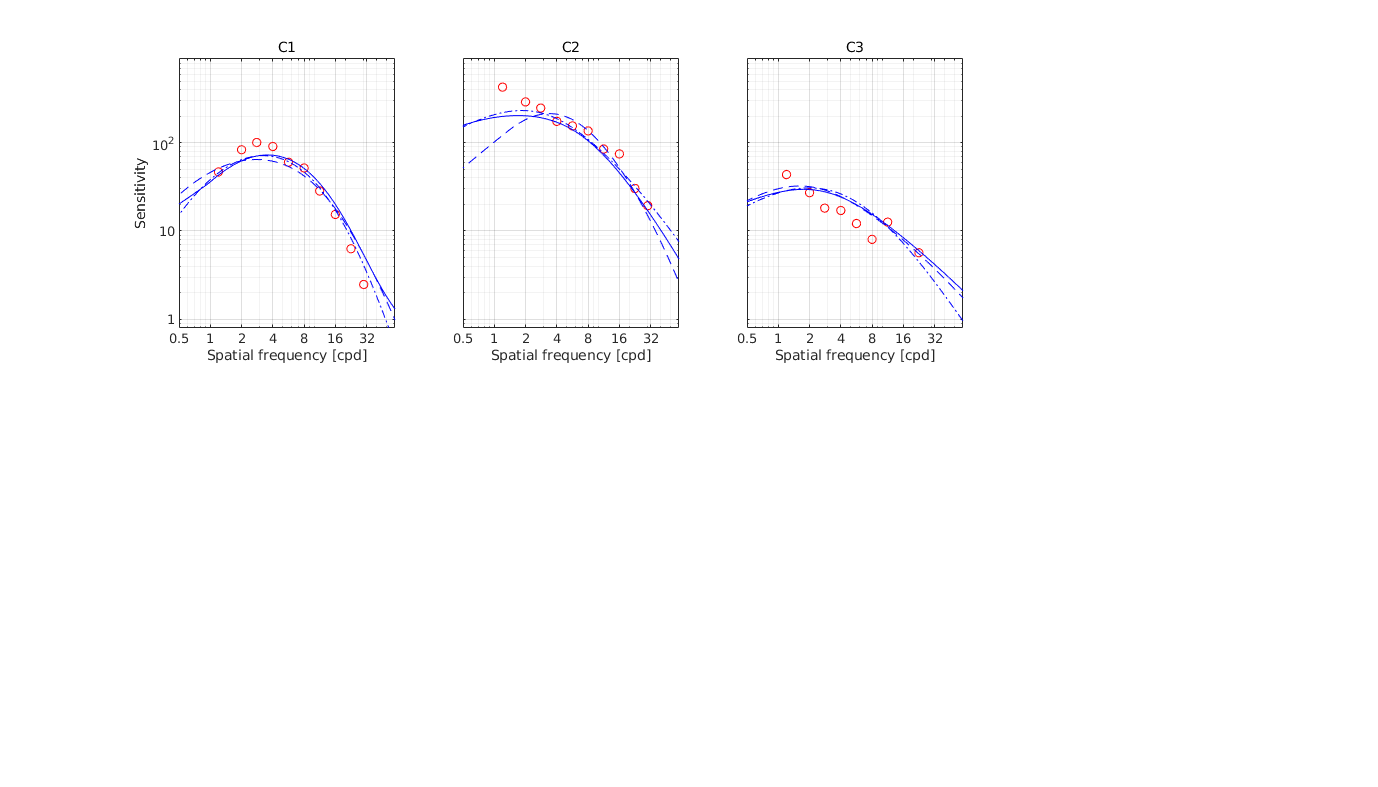
↸Dataset: [colorfest] ColorFest
Chromatic CSF as a function of frequency
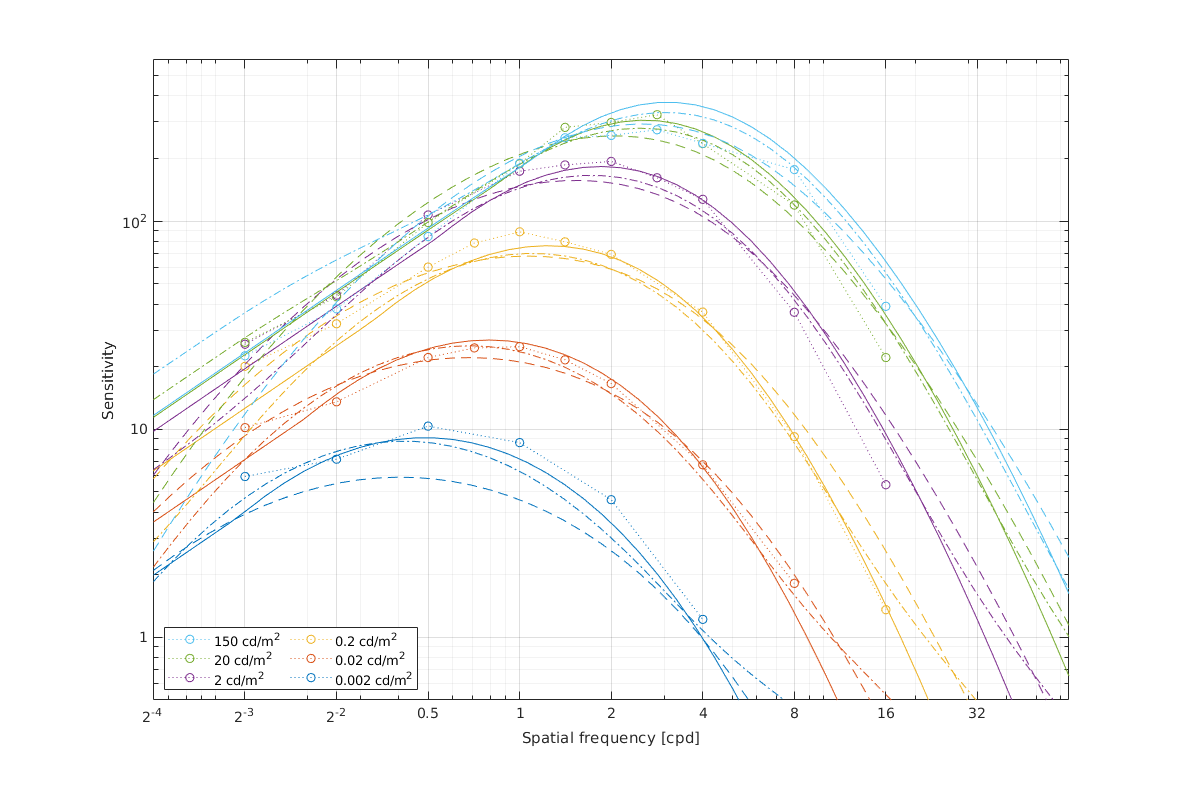
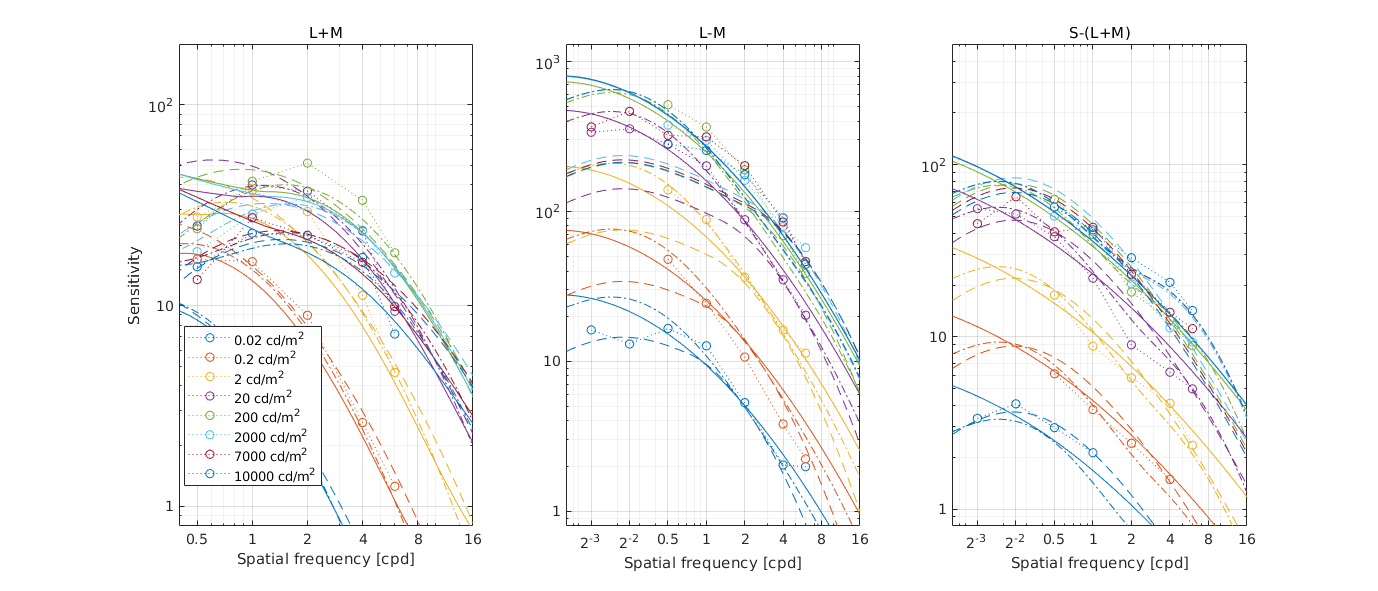
↸Dataset: [hdr_csf] High Dynamic Range CSF
CSF as the function of frequency at different luminance levels (fixed number of cycles)
↸Dataset: [kim2013] Chromatic CSF [Kim et al. 2013]
Chromatic CSF as a function of frequency
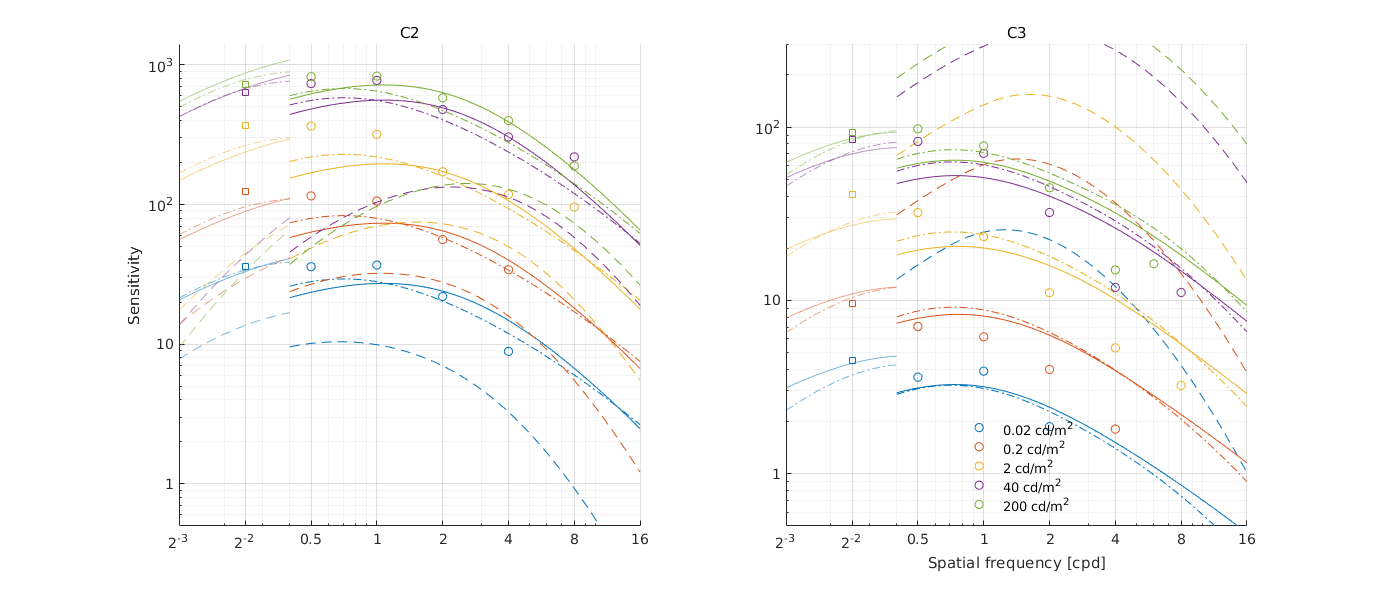
↸Dataset: [hdr_csf_disc] High Dynamic Range Disc CSF
CSF as the function of size at different luminance levels