
Here are my project suggestions for Part II or Masters students. Some of the information on past years' suggestions may also be relevant. I have supervised over 50 Part II, Part III, MPhil and Diploma projects in previous years (see below), with several being singled out for special commendation by the examiners. I also co-authored several academic papers together with former project students of mine. In short, my project suggestions are likely to be challenging but I am fully committed to putting in a lot of effort to provide the best support I can to make sure the project is completed successfully. Who knows, you might end up having a lot of fun too!
Some of the following descriptions are still a bit brief and open to interpretation, watch this space or (better) contact me to find out more. I may add or change project suggestions as the various deadlines approach. Some of the suggestions are based upon projects I have supervised in the past.

Supervisors: Dr Christopher Town and Dr Emily Mitchell
This project concerns recognition of sponge and coral species from modern deep-sea communities. There is a really substantial set of video data created by NOAA (National Oceanic and Atmospheric Administration) and for each data set the different species have been identified along with a dataset of the different species in different environments. The videos are each a few hours long (it varies), but often there are key communities within them that are good to focus on. For example:
https://www.ncei.noaa.gov/waf/okeanos-rov-cruises/ex1706/#tab-11
The main interface for the NOAA deep-sea sponge and coral project includes a searchable database with photographs.
Species IDs are found here, and I would recommend starting with family and maybe genus level.
Details on training data: https://arxiv.org/abs/2007.00114
Here are some papers that look promising as a starting point:
Deep Learning on Underwater Marine Object Detection: A Survey
Deep Learning for Coral Classification
CoralSeg: Learning coral segmentation from sparse annotations
Evaluation of Transfer Learning Scenariosin Plankton Image Classification
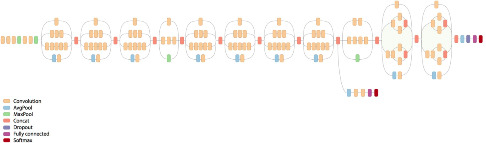
I have in the past done work on using SIFT and other keypoint features to match biological patterns, which can be used for tasks such as the automated identification of individual animals for species at threat of extinction (e.g. manta rays, various sharks, whales, turtles, lizards etc). This project will compare the discriminative capabilities of standard keypoint features (SIFT, SURF, ORB etc) with those derived by modern deep learning architectures that have been pre-trained on large scale image classification tasks.
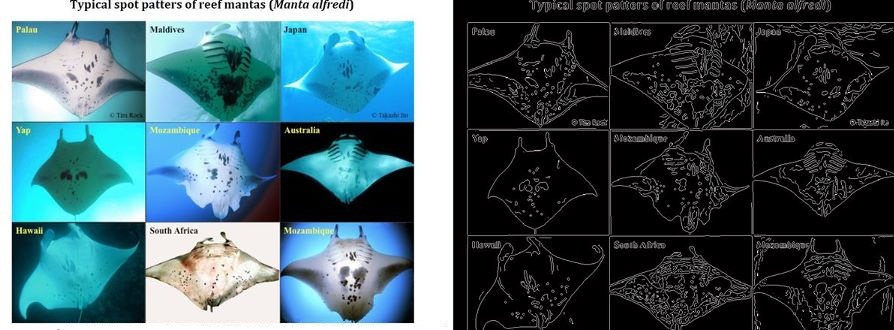
While technologies such as tagging are being used to track and thereby learn more about the behaviour of migratory megafauna, most field work continues to rely on visual identification to catalog and recognise particular specimens. The problems of visual identification are exacerbated in the case of large pelagic marine animals such as whales, manta rays, and sharks, many of whom exhibit characteristic markings that in principle allow individuals to be identified. To this day very little is understood about the roaming behaviour of many of the most magnificent animals in our oceans, and marine scientists and conservationists are having to deal with the task of matching observations against databases of hundreds or thousands of photographs, often taken thousands of miles apart and under very different conditions (lighting, visibility, viewing angle, distance from the subject, occlusions etc.).
This project will build on existing research into using computer vision techniques to extract, analyse, and match characteristic visual patterns in large marine animals. One such system has already been developed by me and has been deployed for research into manta rays, see Manta Matcher.
While the main goal will be to produce a good dissertation, it would of course be desirable to take account of the needs of marine scientists working on projects such as manta ray ecology (see also this page) and the Foundation for the Protection of Marine Megafauna in order to make a contribution towards tools such as the ECOCEAN Whale Shark Photo-identification Library.

Supervisors: Dr Christopher Town and Dr Emily Mitchell
The oldest known animal fossil are found during the Ediacaran time period, around 580 million years ago. These Ediacaran organisms have body-plans unique to the Ediacaran - they are not found elsewhere in the fossil record or alive today. However, the fossil preservation is exceptional, with entire communities of soft-bodied, immobile organisms preserved containing thousands of organisms preserved in-situ across hundreds of rock surfaces in Canada, UK, Australia and Russia. Spatial analyses of these communities can provide a detailed view of Ediacaran life because organisms are preserved in-situ resulting in the near-census preservation of the entire community, with detailed statistical analysis of the positions of the fossils on the bedding plane enabling the rigorous testing of hypotheses relating to Ediacaran life. This spatial approach is still in its infancy, with only a very small proportion of Ediacaran communities currently mapped. This current lack of data is due in part to the enormously time-consuming task of documenting all fossils in the field. However, the use of a cutting edge hand-held laser scanner enables 3D maps of fossil surfaces to be recorded quickly on-site. The laser scans have a sufficiently high resolution (~50 microns) that specimens can be accurately identified away from the locality, thus greatly expanding the number of communities recorded. Once mapped, these fossil spatial distributions can be analysed using spatial point process analyses (SPPA) to describe the spatial patterns present and determine the most likely underlying ecological and biological processes.
This project will investigate whether it is possible to identify where fossils are within the laser scans and resolve the species of the fossils. The data is in three formats: point clouds (raw data), meshes (stl) and images. This project has a few difficulties compared to image recognition on living taxa.
1) The fossil are very low relief which means that on-site they can normally only be spotted with the correct type of angled light, so that photographs are of limited use.
2) Different rock surfaces have different types of fossil preservation, so that recognising species across surfaces can be hard.
3) All the characters of the fossil organism may not have been preserved, or parts of the organism may have been distorted.
Some 3D mesh files can be found here: https://sketchfab.com/egmitchell/collections/ediacaran-fossils
References
Xiao S, Laflamme M. 2009. On the eve of animal radiation: phylogeny, ecology and evolution of the Ediacara biota. Trends in Ecology & Evolution 24: 31-40.
Mitchell EG, Kenchington CG, Liu AG, Matthews JJ, Butterfield NJ. 2015. Reconstructing the reproductive mode of an Ediacaran macro-organism. Nature 524: 343-346.
Mitchell, E.G. and Kenchington, C.G., 2018. The utility of height for the Ediacaran organisms of Mistaken Point. Nature Ecology and Evolution, 2, pp.1218-1222.
Part II, Part III, MPhil, and Diploma in Computer Science student projects at the University of Cambridge (co)supervised by Chris Town